Medical — Publications
Reducing Large Language Model Safety Risks in Women's Health using Semantic Entropy
Large language models (LLMs) hold substantial promise for clinical decision support. However, their widespread adoption in medicine, particularly in healthcare, is hindered by their propensity to generate false or misleading outputs, known as hallucinations. In high-stakes domains such as women’s health (obstetrics & gynaecology), where errors in clinical reasoning can have profound consequences for maternal and neonatal outcomes, ensuring the reliability of AI-generated responses is critical. Traditional methods for quantifying uncertainty, such as perplexity, fail to capture meaning-level inconsistencies that lead to misinformation. Here, we evaluate semantic entropy (SE), a novel uncertainty metric that assesses meaning-level variation, to detect hallucinations in AI-generated medical content. Using a clinically validated dataset derived from UK RCOG MRCOG examinations, we compared SE with perplexity in identifying uncertain responses. SE demonstrated superior performance... [full abstract]
Jahan C. Penny-Dimri, Magdalena Bachmann, William R. Cooke, Sam Mathewlynn, Samual Dockree, John Tolladay, Jannik Kossen, Lin Li, Yarin Gal, Gabriel Jones
arXiv
[paper]
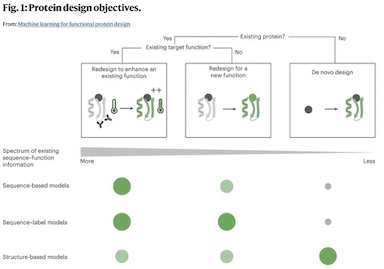
Machine learning for functional protein design
Recent breakthroughs in AI coupled with the rapid accumulation of protein sequence and structure data have radically transformed computational protein design. New methods promise to escape the constraints of natural and laboratory evolution, accelerating the generation of proteins for applications in biotechnology and medicine. To make sense of the exploding diversity of machine learning approaches, we introduce a unifying framework that classifies models on the basis of their use of three core data modalities: sequences, structures and functional labels. We discuss the new capabilities and outstanding challenges for the practical design of enzymes, antibodies, vaccines, nanomachines and more. We then highlight trends shaping the future of this field, from large-scale assays to more robust benchmarks, multimodal foundation models, enhanced sampling strategies and laboratory automation.
Pascal Notin, Nathan Rollins, Yarin Gal, Chris Sander, Debora Marks
Nature Biotechnology (2024)
[paper]
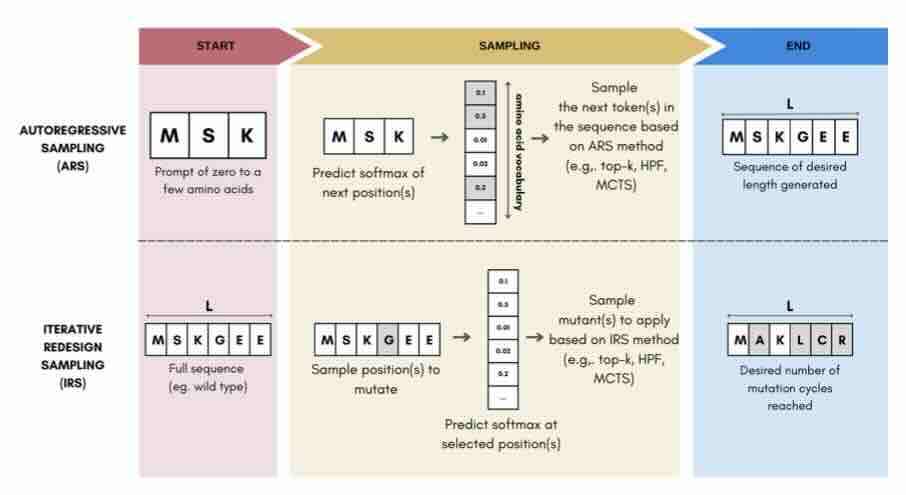
Sampling Protein Language Models for Functional Protein Design
Protein language models have emerged as powerful ways to learn complex repre- sentations of proteins, thereby improving their performance on several downstream tasks, from structure prediction to fitness prediction, property prediction, homology detection, and more. By learning a distribution over protein sequences, they are also very promising tools for designing novel and functional proteins, with broad applications in healthcare, new material, or sustainability. Given the vastness of the corresponding sample space, efficient exploration methods are critical to the success of protein engineering efforts. However, the methodologies for ade- quately sampling these models to achieve core protein design objectives remain underexplored and have predominantly leaned on techniques developed for Natural Language Processing. In this work, we first develop a holistic in silico protein design evaluation framework, to comprehensively compare different sampling methods. After performing a ... [full abstract]
Jeremie Theddy Darmawan, Yarin Gal, Pascal Notin
Machine Learning for Structural Biology / Generative AI and Biology workshops, NeurIPS 2023
[Paper]
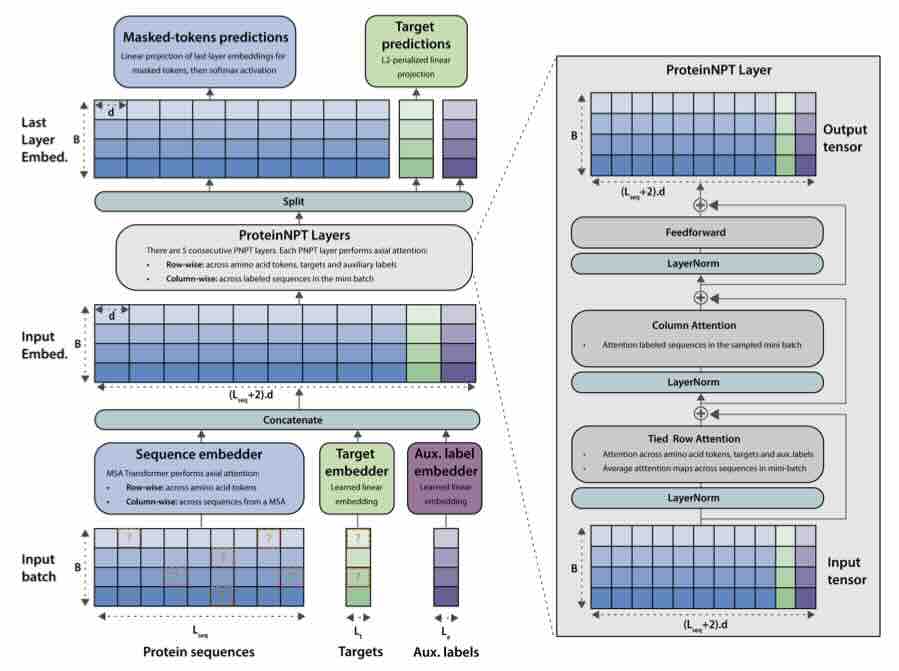
ProteinNPT: Improving Protein Property Prediction and Design with Non-Parametric Transformers
Protein design holds immense potential for optimizing naturally occurring proteins, with broad applications in drug discovery, material design, and sustainability. How- ever, computational methods for protein engineering are confronted with significant challenges, such as an expansive design space, sparse functional regions, and a scarcity of available labels. These issues are further exacerbated in practice by the fact most real-life design scenarios necessitate the simultaneous optimization of multiple properties. In this work, we introduce ProteinNPT, a non-parametric trans- former variant tailored to protein sequences and particularly suited to label-scarce and multi-task learning settings. We first focus on the supervised fitness prediction setting and develop several cross-validation schemes which support robust perfor- mance assessment. We subsequently reimplement prior top-performing baselines, introduce several extensions of these baselines by integrating diverse branch... [full abstract]
Pascal Notin, Ruben Weitzman, Debora Marks, Yarin Gal
NeurIPS 2023
[Paper]
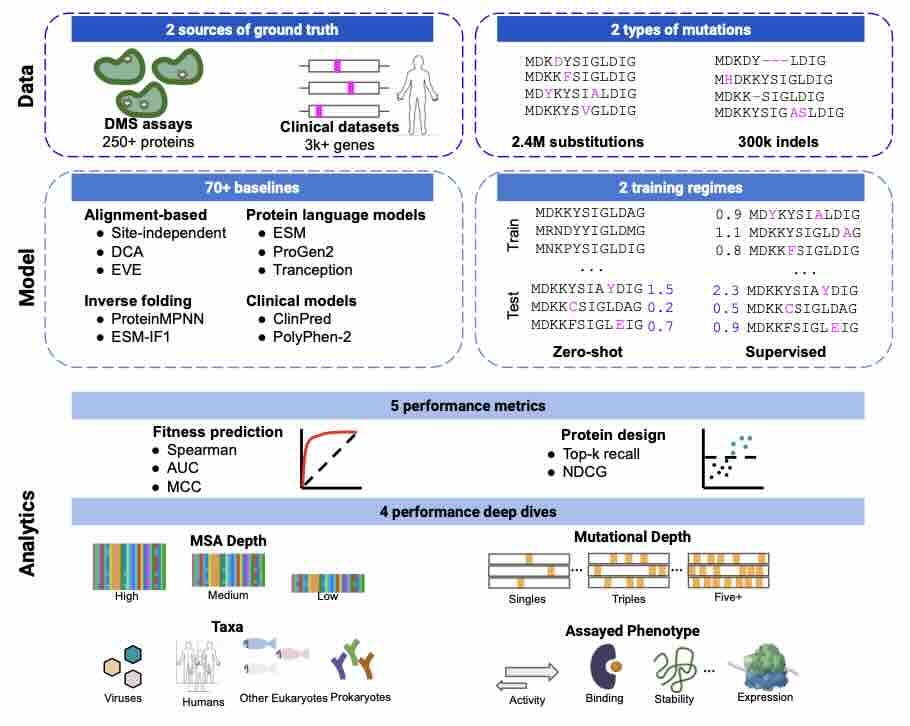
ProteinGym: Large-Scale Benchmarks for Protein Fitness Prediction and Design
Predicting the effects of mutations in proteins is critical to many applications, from understanding genetic disease to designing novel proteins that can address our most pressing challenges in climate, agriculture and healthcare. Despite a surge in machine learning-based protein models to tackle these questions, an assessment of their respective benefits is challenging due to the use of distinct, often contrived, experimental datasets, and the variable performance of models across different protein families. Addressing these challenges requires scale. To that end we introduce ProteinGym, a large-scale and holistic set of benchmarks specifically designed for protein fitness prediction and design. It encompasses both a broad collection of over 250 standardized deep mutational scanning assays, spanning millions of mutated sequences, as well as curated clinical datasets providing high- quality expert annotations about mutation effects. We devise a robust evaluation framework that c... [full abstract]
Pascal Notin, Aaron W. Kollasch, Daniel Ritter, Lood van Niekerk, Steffanie Paul, Hansen Spinner, Nathan Rollins, Ada Shaw, Ruben Weitzman, Jonathan Frazer, Mafalda Dias, Dinko Franceschi, Rose Orenbuch, Yarin Gal, Debora Marks
NeurIPS 2023
[Paper]
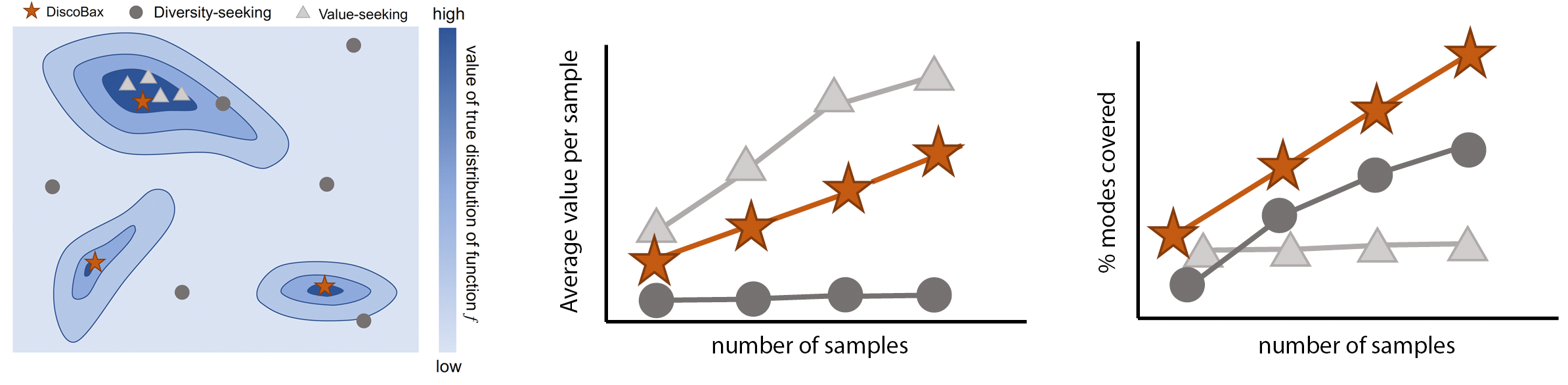
DiscoBAX - Discovery of optimal intervention sets in genomic experiment design
The discovery of novel therapeutics to cure genetic pathologies relies on the identification of the different genes involved in the underlying disease mechanism. With billions of potential hypotheses to test, an exhaustive exploration of the entire space of potential interventions is impossible in practice. Sample-efficient methods based on active learning or bayesian optimization bear the promise of identifying interesting targets using the least experiments possible. However, genomic perturbation experiments typically rely on proxy outcomes measured in biological model systems that may not completely correlate with the outcome of interventions in humans. In practical experiment design, one aims to find a set of interventions which maximally move a target phenotype via a diverse set of mechanisms in order to reduce the risk of failure in future stages of trials. To that end, we introduce DiscoBAX — a sample-efficient algorithm for the discovery of genetic interventions that ma... [full abstract]
Clare Lyle, Arash Mehrjou, Pascal Notin, Andrew Jesson, Stefan Bauer, Yarin Gal, Patrick Schwab
ICML 2023
[arXiv]
Mixtures of large-scale dynamic functional brain network modes
Accurate temporal modelling of functional brain networks is essential in the quest for understanding how such networks facilitate cognition. Researchers are beginning to adopt time-varying analyses for electrophysiological data that capture highly dynamic processes on the order of milliseconds. Typically, these approaches, such as clustering of functional connectivity profiles and Hidden Markov Modelling (HMM), assume mutual exclusivity of networks over time. Whilst a powerful constraint, this assumption may be compromising the ability of these approaches to describe the data effectively. Here, we propose a new generative model for functional connectivity as a time-varying linear mixture of spatially distributed statistical “modes”. The temporal evolution of this mixture is governed by a recurrent neural network, which enables the model to generate data with a rich temporal structure. We use a Bayesian framework known as amortised variational inference to learn model parameters ... [full abstract]
Chetan Gohil, Evan Roberts, Ryan Timms, Alex Skates, Cameron Higgins, Andrew Quinn, Usama Pervaiz, Joost van Amersfoort, Pascal Notin, Yarin Gal, Stanislaw Adaszewski, Mark Woolrich
NeuroImage
[paper]
Seasonal variation in SARS-CoV-2 transmission in temperate climates: A Bayesian modelling study in 143 European regions
Although seasonal variation has a known influence on the transmission of several respiratory viral infections, its role in SARS-CoV-2 transmission remains unclear. While there is a sizable and growing literature on environmental drivers of COVID-19 transmission, recent reviews have highlighted conflicting and inconclusive findings. This indeterminacy partly owes to the fact that seasonal variation relates to viral transmission by a complicated web of causal pathways, including many interacting biological and behavioural factors. Since analyses of specific factors cannot determine the aggregate strength of seasonal forcing, we sidestep the challenge of disentangling various possible causal paths in favor of a holistic approach. We model seasonality as a sinusoidal variation in transmission and infer a single Bayesian estimate of the overall seasonal effect. By extending two state-of-the-art models of non-pharmaceutical intervention (NPI) effects and their datasets covering 143 re... [full abstract]
Tomáš Gavenčiak*, Joshua Teperowsky Monrad*, Gavin Leech, Mrinank Sharma, Sören Mindermann, Samir Bhatt, Jan Brauner, Jan Kulveit*
PLoS Computational Biology 18(8): e1010435. (2022)
[Paper]
Plex: Towards Reliability using Pretrained Large Model Extensions
A recent trend in artificial intelligence is the use of pretrained models for language and vision tasks, which have achieved extraordinary performance but also puzzling failures. Probing these models’ abilities in diverse ways is therefore critical to the field. In this paper, we explore the reliability of models, where we define a reliable model as one that not only achieves strong predictive performance but also performs well consistently over many decision-making tasks involving uncertainty (e.g., selective prediction, open set recognition), robust generalization (e.g., accuracy and proper scoring rules such as log-likelihood on in- and out-of-distribution datasets), and adaptation (e.g., active learning, few-shot uncertainty). We devise 10 types of tasks over 40 datasets in order to evaluate different aspects of reliability on both vision and language domains. To improve reliability, we developed ViT-Plex and T5-Plex, pretrained large model extensions for vision and language... [full abstract]
Dustin Tran, Jeremiah Liu, Michael W. Dusenberry, Du Phan, Mark Collier, Jie Ren, Kehang Han, Zi Wang, Zelda Mariet, Huiyi Hu, Neil Band, Tim G. J. Rudner, Karan Singhal, Zachary Nado, Joost van Amersfoort, Andreas Kirsch, Rodolphe Jenatton, Nithum Thain, Honglin Yuan, Kelly Buchanan, Kevin Murphy, D. Sculley, Yarin Gal, Zoubin Ghahramani, Jasper Snoek, Balaji Lakshminarayan
Contributed Talk, ICML Pre-training Workshop, 2022
[OpenReview] [Code] [BibTex] [Google AI Blog Post]
Learning from pre-pandemic data to forecast viral antibody escape
From early detection of variants of concern to vaccine and therapeutic design, pandemic preparedness depends on identifying viral mutations that escape the response of the host immune system. While experimental scans are useful for quantifying escape potential, they remain laborious and impractical for exploring the combinatorial space of mutations. Here we introduce a biologically grounded model to quantify the viral escape potential of mutations at scale. Our method - EVEscape - brings together fitness predictions from evolutionary models, structure-based features that assess antibody binding potential, and distances between mutated and wild-type residues. Unlike other models that predict variants of concern based on newly observed variants, EVEscape has no reliance on recent community prevalence, and is applicable before surveillance sequencing or experimental scans are broadly available. We validate EVEscape predictions against experimental data on H1N1, HIV and SARS-CoV-2, ... [full abstract]
Nicole N Thadani, Sarah Gurev, Pascal Notin, Noor Youssef, Nathan J Rollins, Daniel Ritter, Chris Sander, Yarin Gal, Debora Marks
Nature
[paper]
Tranception: protein fitness prediction with autoregressive transformers and inference-time retrieval
The ability to accurately model the fitness landscape of protein sequences is critical to a wide range of applications, from quantifying the effects of human variants on disease likelihood, to predicting immune-escape mutations in viruses and designing novel biotherapeutic proteins. Deep generative models of protein sequences trained on multiple sequence alignments have been the most successful approaches so far to address these tasks. The performance of these methods is however contingent on the availability of sufficiently deep and diverse alignments for reliable training. Their potential scope is thus limited by the fact many protein families are hard, if not impossible, to align. Large language models trained on massive quantities of non-aligned protein sequences from diverse families address these problems and show potential to eventually bridge the performance gap. We introduce Tranception, a novel transformer architecture leveraging autoregressive predictions and retrieva... [full abstract]
Pascal Notin, Mafalda Dias, Jonathan Frazer, Javier Marchena-Hurtado, Aidan Gomez, Debora Marks, Yarin Gal
ICML, 2022
[Preprint] [BibTex] [Code]
Mask wearing in community settings reduces SARS-CoV-2 transmission
The effectiveness of mask wearing at controlling severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) transmission has been unclear. While masks are known to substantially reduce disease transmission in healthcare settings, studies in community settings report inconsistent results. Most such studies focus on how masks impact transmission, by analyzing how effective government mask mandates are. However, we find that widespread voluntary mask wearing, and other data limitations, make mandate effectiveness a poor proxy for mask-wearing effectiveness. We directly analyze the effect of mask wearing on SARS-CoV-2 transmission, drawing on several datasets covering 92 regions on six continents, including the largest survey of wearing behavior (n= 20 million). Using a Bayesian hierarchical model, we estimate the effect of mask wearing on transmission, by linking reported wearing levels to reported cases in each region, while adjusting for mobility and nonpharmaceutical intervent... [full abstract]
Gavin Leech, Charlie Rogers-Smith, Joshua Teperowsky Monrad, Jonas B. Sandbrink, Benedict Snodin, Robert Zinkov, Benjamin Rader, John S. Brownstein, Yarin Gal, Samir Bhatt, Mrinank Sharma, Sören Mindermann, Jan Brauner, Laurence Aitchinson
Proceedings of the National Academy of Sciences (PNAS) (2022) 119 (23) e2119266119
[Paper]
RITA: a Study on Scaling Up Generative Protein Sequence Models
In this work we introduce RITA: a suite of autoregressive generative models for protein sequences, with up to 1.2 billion parameters, trained on over 280 million protein sequences belonging to the UniRef-100 database. Such generative models hold the promise of greatly accelerating protein design. We conduct the first systematic study of how capabilities evolve with model size for autoregressive transformers in the protein domain: we evaluate RITA models in next amino acid prediction, zero-shot fitness, and enzyme function prediction, showing benefits from increased scale. We release the RITA models openly, to the benefit of the research community.
Daniel Hesslow, Niccoló Zanichelli, Pascal Notin, Iacopo Poli, Debora Marks
ICML, Workshop on Computational biology, 2022 (Spotlight)
[Preprint] [Code] [BibTex]
GeneDisco: A Benchmark for Experimental Design in Drug Discovery
In vitro cellular experimentation with genetic interventions, using for example CRISPR technologies, is an essential step in early-stage drug discovery and target validation that serves to assess initial hypotheses about causal associations between biological mechanisms and disease pathologies. With billions of potential hypotheses to test, the experimental design space for in vitro genetic experiments is extremely vast, and the available experimental capacity - even at the largest research institutions in the world - pales in relation to the size of this biological hypothesis space. Machine learning methods, such as active and reinforcement learning, could aid in optimally exploring the vast biological space by integrating prior knowledge from various information sources as well as extrapolating to yet unexplored areas of the experimental design space based on available data. However, there exist no standardised benchmarks and data sets for this challenging task and little rese... [full abstract]
Arash Mehrjou, Ashkan Soleymani, Andrew Jesson, Pascal Notin, Yarin Gal, Stefan Bauer, Patrick Schwab
International Conference on Learning Representations, 2022
[Preprint] [BibTex] [Code]
Mixtures of large-scale dynamic functional brain network modes
Accurate temporal modelling of functional brain networks is essential in the quest for understanding how such networks facilitate cognition. Researchers are beginning to adopt time-varying analyses for electrophysiological data that capture highly dynamic processes on the order of milliseconds. Typically, these approaches, such as clustering of functional connectivity profiles and Hidden Markov Modelling (HMM), assume mutual exclusivity of networks over time. Whilst a powerful constraint, this assumption may be compromising the ability of these approaches to describe the data effectively. Here, we propose a new generative model for functional connectivity as a time-varying linear mixture of spatially distributed statistical “modes”. The temporal evolution of this mixture is governed by a recurrent neural network, which enables the model to generate data with a rich temporal structure. We use a Bayesian framework known as amortised variational inference to learn model parameters ... [full abstract]
Chetan Gohil, Evan Roberts, Ryan Timms, Alex Skates, Cameron Higgins, Andrew Quinn, Usama Pervaiz, Joost van Amersfoort, Pascal Notin, Yarin Gal, Stanislaw Adaszewski, Mark Woolrich
NeuroImage Volume 263 [Paper] [BibTex]
Understanding the effectiveness of government interventions against the resurgence of COVID-19 in Europe
During the second half of 2020, many European governments responded to the resurging transmission of SARS-CoV-2 with wide-ranging non-pharmaceutical interventions (NPIs). These efforts were often highly targeted at the regional level and included fine-grained NPIs. This paper describes a new dataset designed for the accurate recording of NPIs in Europe’s second wave to allow precise modelling of NPI effectiveness. The dataset includes interventions from 114 regions in 7 European countries during the period from the 1st August 2020 to the 9th January 2021. The paper includes NPI definitions tailored to the second wave following an exploratory data collection. Each entry has been extensively validated by semi-independent double entry, comparison with existing datasets, and, when necessary, discussion with local epidemiologists. The dataset has considerable potential for use in disentangling the effectiveness of NPIs and comparing the impact of interventions across different phases... [full abstract]
George Altman, Janvi Ahuja, Joshua Teperowsky Monrad, Gurpreet Dhaliwal, Charlie Rogers-Smith, Gavin Leech, Benedict Snodin, Jonas B. Sandbrink, Lukas Finnveden, Alexander John Norman, Sebastian B. Oehm, Julia Fabienne Sandkühler, Jan Kulveit, Seth Flaxman, Yarin Gal, Swapnil Mishra, Samir Bhatt, Mrinank Sharma, Sören Mindermann, Jan Brauner
Nature Scientific Data 9, Article number: 145 (2022)
[Paper]
All-cause versus cause-specific excess deaths for estimating influenza-associated mortality in Denmark, Spain, and the United States
Seasonal influenza-associated excess mortality estimates can be timely and provide useful information on the severity of an epidemic. This methodology can be leveraged during an emergency response or pandemic. For Denmark, Spain, and the United States, we estimated age-stratified excess mortality for (i) all-cause, (ii) respiratory and circulatory, (iii) circulatory, (iv) respiratory, and (v) pneumonia, and influenza causes of death for the 2015/2016 and 2016/2017 influenza seasons. We quantified differences between the countries and seasonal excess mortality estimates and the death categories. We used a time-series linear regression model accounting for time and seasonal trends using mortality data from 2010 through 2017. The respective periods of weekly excess mortality for all-cause and cause-specific deaths were similar in their chronological patterns. Seasonal all-cause excess mortality rates for the 2015/2016 and 2016/2017 influenza seasons were 4.7 (3.3–6.1) and 14.3 (13.... [full abstract]
Sebastian SS Schmidt, Angela Danielle Iuliano, Lasse S Vestergaard, Clara Mazagatos‐Ateca, Amparo Larrauri, Jan Brauner, Sonja J Olsen, Jens Nielsen, Joshua A Salomon, Tyra G Krause
Influenza and Other Respiratory Viruses, Volume 16, Issue 4
[paper]
QU-BraTS: MICCAI BraTS 2020 Challenge on Quantifying Uncertainty in Brain Tumor Segmentation -- Analysis of Ranking Metrics and Benchmarking Results
Deep learning (DL) models have provided the state-of-the-art performance in a wide variety of medical imaging benchmarking challenges, including the Brain Tumor Segmentation (BraTS) challenges. However, the task of focal pathology multi-compartment segmentation (e.g., tumor and lesion sub-regions) is particularly challenging, and potential errors hinder the translation of DL models into clinical workflows. Quantifying the reliability of DL model predictions in the form of uncertainties, could enable clinical review of the most uncertain regions, thereby building trust and paving the way towards clinical translation. Recently, a number of uncertainty estimation methods have been introduced for DL medical image segmentation tasks. Developing metrics to evaluate and compare the performance of uncertainty measures will assist the end-user in making more informed decisions. In this study, we explore and evaluate a metric developed during the BraTS 2019-2020 task on uncertainty quanti... [full abstract]
Raghav Mehta, Angelos Filos, Spyridon Bakas, Yarin Gal, Tal Arbel
Preprint (19 Dec 2021)
[Paper]
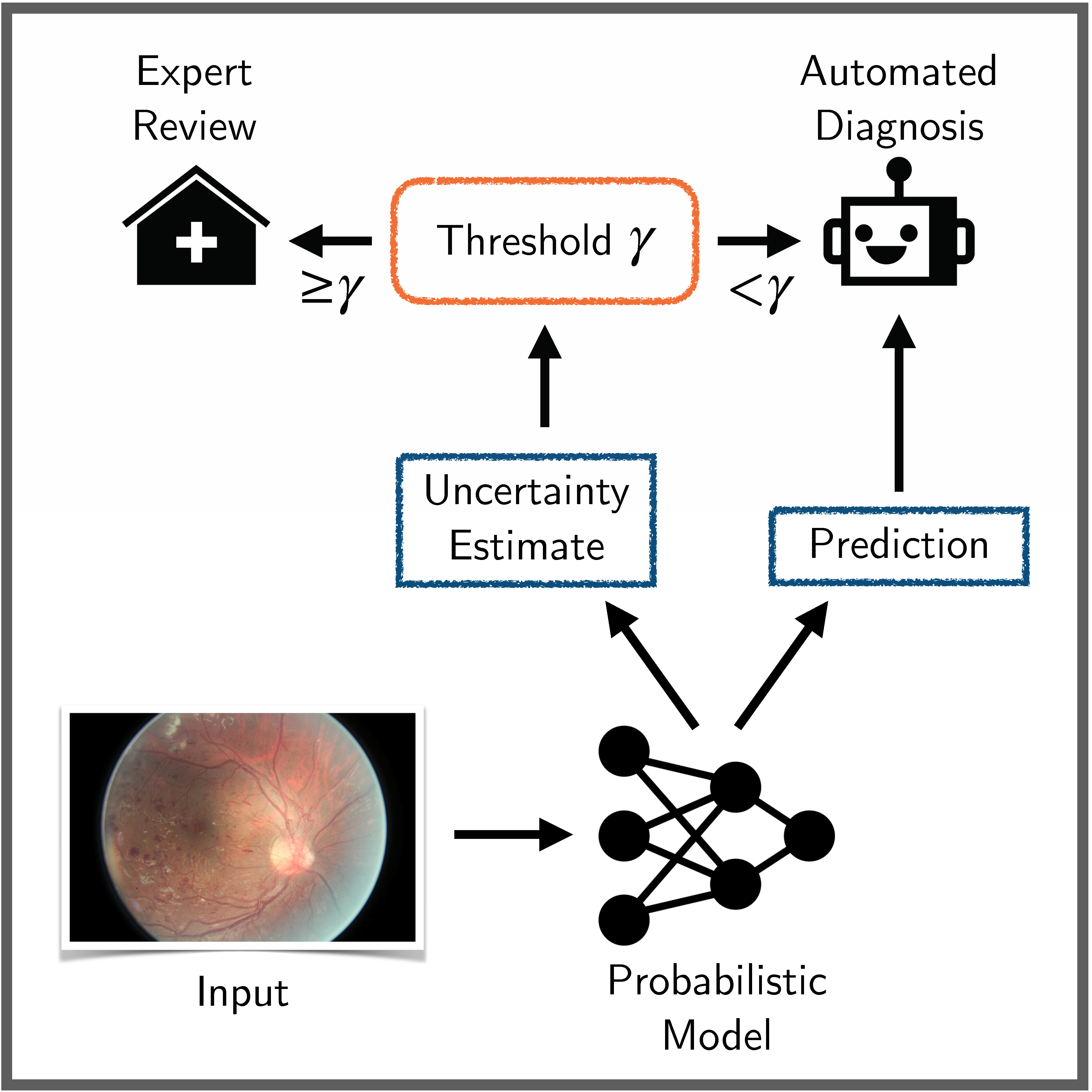
Benchmarking Bayesian Deep Learning on Diabetic Retinopathy Detection Tasks
Bayesian deep learning seeks to equip deep neural networks with the ability to precisely quantify their predictive uncertainty, and has promised to make deep learning more reliable for safety-critical real-world applications. Yet, existing Bayesian deep learning methods fall short of this promise; new methods continue to be evaluated on unrealistic test beds that do not reflect the complexities of downstream real-world tasks that would benefit most from reliable uncertainty quantification. We propose a set of real-world tasks that accurately reflect such complexities and are designed to assess the reliability of predictive models in safety-critical scenarios. Specifically, we curate two publicly available datasets of high-resolution human retina images exhibiting varying degrees of diabetic retinopathy, a medical condition that can lead to blindness, and use them to design a suite of automated diagnosis tasks that require reliable predictive uncertainty quantification. We use th... [full abstract]
Neil Band, Tim G. J. Rudner, Qixuan Feng, Angelos Filos, Zachary Nado, Michael W. Dusenberry, Ghassen Jerfel, Dustin Tran, Yarin Gal
NeurIPS Datasets and Benchmarks Track, 2021
Spotlight Talk, NeurIPS Workshop on Distribution Shifts, 2021
Symposium on Machine Learning for Health (ML4H) Extended Abstract Track, 2021
NeurIPS Workshop on Bayesian Deep Learning, 2021
[OpenReview] [Code] [BibTex]
Uncertainty Baselines: Benchmarks for Uncertainty & Robustness in Deep Learning
High-quality estimates of uncertainty and robustness are crucial for numerous real-world applications, especially for deep learning which underlies many deployed ML systems. The ability to compare techniques for improving these estimates is therefore very important for research and practice alike. Yet, competitive comparisons of methods are often lacking due to a range of reasons, including: compute availability for extensive tuning, incorporation of sufficiently many baselines, and concrete documentation for reproducibility. In this paper we introduce Uncertainty Baselines: high-quality implementations of standard and state-of-the-art deep learning methods on a variety of tasks. As of this writing, the collection spans 19 methods across 9 tasks, each with at least 5 metrics. Each baseline is a self-contained experiment pipeline with easily reusable and extendable components. Our goal is to provide immediate starting points for experimentation with new methods or applications. A... [full abstract]
Zachary Nado, Neil Band, Mark Collier, Josip Djolonga, Michael W. Dusenberry, Sebastian Farquhar, Angelos Filos, Marton Havasi, Rodolphe Jenatton, Ghassen Jerfel, Jeremiah Liu, Zelda Mariet, Jeremy Nixon, Shreyas Padhy, Jie Ren, Tim G. J. Rudner, Yeming Wen, Florian Wenzel, Kevin Murphy, D. Sculley, Balaji Lakshminarayanan, Jasper Snoek, Yarin Gal, Dustin Tran
NeurIPS Workshop on Bayesian Deep Learning, 2021
[arXiv] [Code] [Blog Post (Google AI)] [BibTex]
Disease variant prediction with deep generative models of evolutionary data
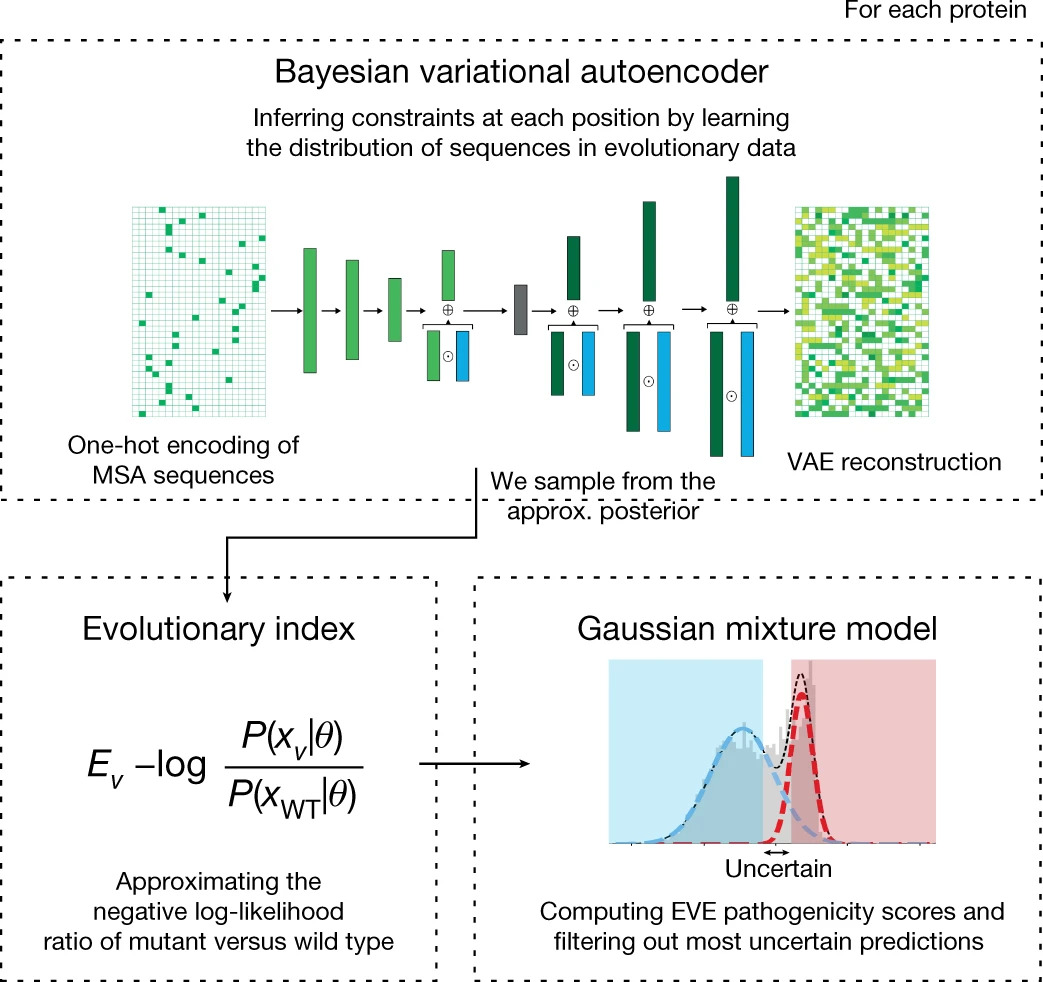
Quantifying the pathogenicity of protein variants in human disease-related genes would have a marked effect on clinical decisions, yet the overwhelming majority (over 98%) of these variants still have unknown consequences. In principle, computational methods could support the large-scale interpretation of genetic variants. However, state-of-the-art methods have relied on training machine learning models on known disease labels. As these labels are sparse, biased and of variable quality, the resulting models have been considered insufficiently reliable. Here we propose an approach that leverages deep generative models to predict variant pathogenicity without relying on labels. By modelling the distribution of sequence variation across organisms, we implicitly capture constraints on the protein sequences that maintain fitness. Our model EVE (evolutionary model of variant effect) not only outperforms computational approaches that rely on labelled data but also performs on par with,... [full abstract]
Jonathan Frazer, Pascal Notin, Mafalda Dias, Aidan Gomez, Joseph K. Min, Kelly Brock, Yarin Gal, Debora Marks
Nature, 2021 (volume 599, pages 91–95)
[Paper] [BibTex] [Preprint] [Website] [Code]
Understanding the effectiveness of government interventions against the resurgence of COVID-19 in Europe
Governments are attempting to control the COVID-19 pandemic with nonpharmaceutical interventions (NPIs). However, the effectiveness of different NPIs at reducing transmission is poorly understood. We gathered chronological data on the implementation of NPIs for several European, and other, countries between January and the end of May 2020. We estimate the effectiveness of NPIs, ranging from limiting gathering sizes, business closures, and closure of educational institutions to stay-at-home orders. To do so, we used a Bayesian hierarchical model that links NPI implementation dates to national case and death counts and supported the results with extensive empirical validation. Closing all educational institutions, limiting gatherings to 10 people or less, and closing face-to-face businesses each reduced transmission considerably. The additional effect of stay-at-home orders was comparatively small.
Mrinank Sharma, Sören Mindermann, Charlie Rogers-Smith, Gavin Leech, Benedict Snodin, Janvi Ahuja, Jonas B. Sandbrink, Joshua Teperowsky Monrad, George Altman, Gurpreet Dhaliwal, Lukas Finnveden, Alexander John Norman, Sebastian B. Oehm, Julia Fabienne Sandkühler, Laurence Aitchison, Tomas Gavenciak, Thomas Mellan, Jan Kulveit, Leonid Chindelevitch, Seth Flaxman, Yarin Gal, Swapnil Mishra, Samir Bhatt, Jan Brauner
Nature Communications (2021) 12: 5820
[Paper]
Causal-BALD: Deep Bayesian Active Learning of Outcomes to Infer Treatment-Effects
Estimating personalized treatment effects from high-dimensional observational data is essential in situations where experimental designs are infeasible, unethical or expensive. Existing approaches rely on fitting deep models on outcomes observed for treated and control populations, but when measuring the outcome for an individual is costly (e.g. biopsy) a sample efficient strategy for acquiring outcomes is required. Deep Bayesian active learning provides a framework for efficient data acquisition by selecting points with high uncertainty. However, naive application of existing methods selects training data that is biased toward regions where the treatment effect cannot be identified because there is non-overlapping support between the treated and control populations. To maximize sample efficiency for learning personalized treatment effects, we introduce new acquisition functions grounded in information theory that bias data acquisition towards regions where overlap is satisfied,... [full abstract]
Andrew Jesson, Panagiotis Tigas, Joost van Amersfoort, Andreas Kirsch, Uri Shalit, Yarin Gal
NeurIPS, 2021
[Paper]
Changing composition of SARS-CoV-2 lineages and rise of Delta variant in England
Background Since its emergence in Autumn 2020, the SARS-CoV-2 Variant of Concern (VOC) B.1.1.7 (WHO label Alpha) rapidly became the dominant lineage across much of Europe. Simultaneously, several other VOCs were identified globally. Unlike B.1.1.7, some of these VOCs possess mutations thought to confer partial immune escape. Understanding when and how these additional VOCs pose a threat in settings where B.1.1.7 is currently dominant is vital.
Methods We examine trends in the prevalence of non-B.1.1.7 lineages in London and other English regions using passive-case detection PCR data, cross-sectional community infection surveys, genomic surveillance, and wastewater monitoring. The study period spans from 31st January 2021 to 15th May 2021.
Findings Across data sources, the percentage of non-B.1.1.7 variants has been increasing since late March 2021. This increase was initially driven by a variety of lineages with immune escape. From mid-April, B.1.617.2 (WHO label... [full abstract]
Swapnil Mishra, Sören Mindermann, Mrinank Sharma, Charles Whittaker, Thomas A. Mellan, Thomas Wilton, Dimitra Klapsa, Ryan Mate, Martin Fritzsche, Maria Zambon, Janvi Ahuja, Adam Howes, Xenia Miscouridou, Guy P. Nason, Oliver Ratmann, Elizaveta Semenova, Gavin Leech, Julia Fabienne Sandkühler, Charlie Rogers-Smith, Michaela Vollmer, H. Juliette T. Unwin, Yarin Gal, Meera Chand, Axel Gandy, Javier Martin, Erik Volz, Neil M. Ferguson, Samir Bhatt, Jan Brauner, Seth Flaxman
EClinicalMedicine (2021), 39:101064
[Paper]
Propagating Uncertainty Across Cascaded Medical Imaging Tasks for Improved Deep Learning Inference
Although deep networks have been shown to perform very well on a variety of medical imaging tasks, inference in the presence of pathology presents several challenges to common models. These challenges impede the integration of deep learning models into real clinical workflows, where the customary process of cascading deterministic outputs from a sequence of image-based inference steps (e.g. registration, segmentation) generally leads to an accumulation of errors that impacts the accuracy of downstream inference tasks. In this paper, we propose that by embedding uncertainty estimates across cascaded inference tasks, performance on the downstream inference tasks should be improved. We demonstrate the effectiveness of the proposed approach in three different clinical contexts: (i) We demonstrate that by propagating T2 weighted lesion segmentation results and their associated uncertainties, subsequent T2 lesion detection performance is improved when evaluated on a proprietary large-... [full abstract]
Raghav Mehta, Thomas Christinck, Tanya Nair, Aurélie Bussy, Swapna Premasiri, Manuela Costantino, M. Mallar Chakravarthy, Douglas L. Arnold, Yarin Gal, Tal Arbel
IEEE Transactions on Medical Imaging, Vol. 41, No. 2 (2022)
[IEEE T-MI]
Improving black-box optimization in VAE latent space using decoder uncertainty
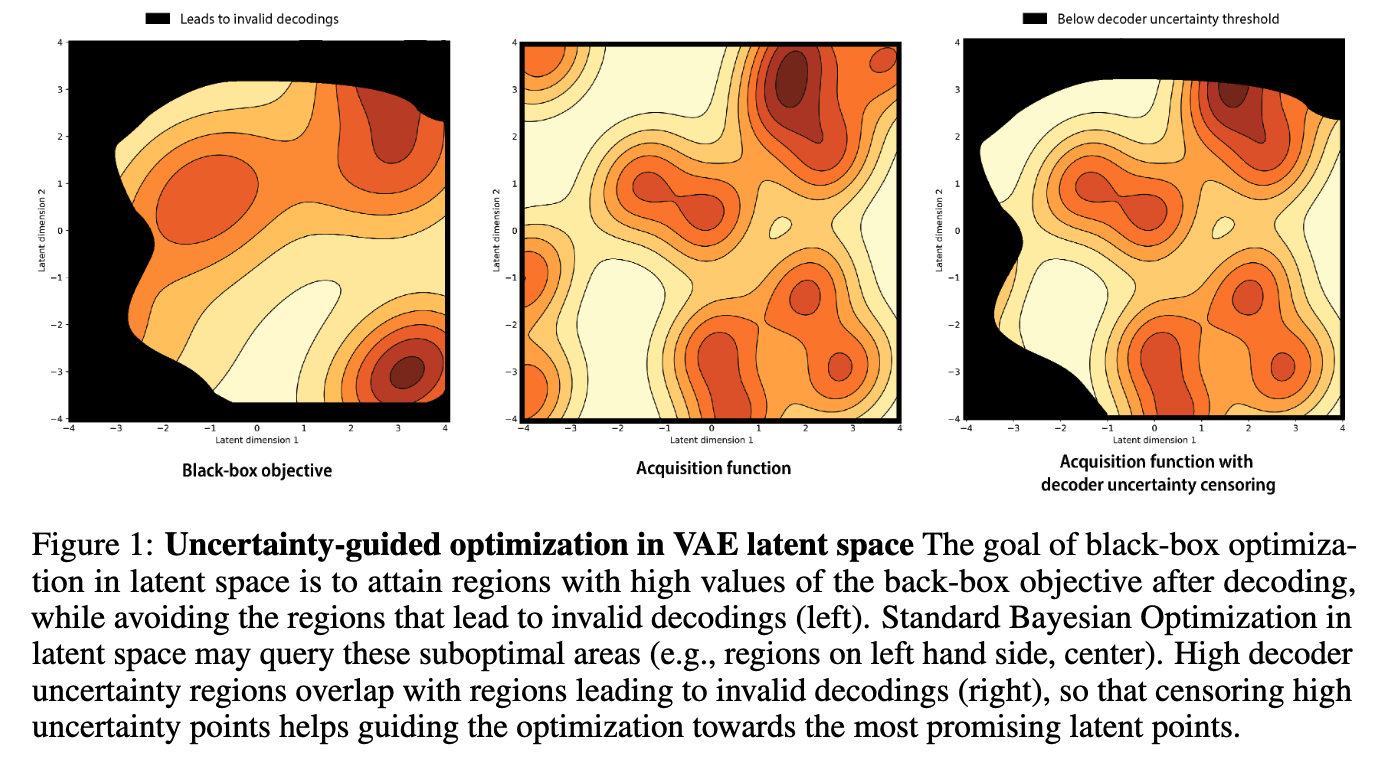
Optimization in the latent space of variational autoencoders is a promising approach to generate high-dimensional discrete objects that maximize an expensive black-box property (e.g., drug-likeness in molecular generation, function approximation with arithmetic expressions). However, existing methods lack robustness as they may decide to explore areas of the latent space for which no data was available during training and where the decoder can be unreliable, leading to the generation of unrealistic or invalid objects. We propose to leverage the epistemic uncertainty of the decoder to guide the optimization process. This is not trivial though, as a naive estimation of uncertainty in the high-dimensional and structured settings we consider would result in high estimator variance. To solve this problem, we introduce an importance sampling-based estimator that provides more robust estimates of epistemic uncertainty. Our uncertainty-guided optimization approach does not require modif... [full abstract]
Pascal Notin, José Miguel Hernández-Lobato, Yarin Gal
NeurIPS, 2021
[Preprint] [Proceedings] [BibTex] [Code]
Is the cure really worse than the disease? The health impacts of lockdowns during COVID-19
During the pandemic, there has been ongoing and contentious debate around the impact of restrictive government measures to contain SARS-CoV-2 outbreaks, often termed ‘lockdowns’. We define a ‘lockdown’ as a highly restrictive set of non-pharmaceutical interventions against COVID-19, including either stay-at-home orders or interventions with an equivalent effect on movement in the population through restriction of movement. While necessarily broad, this definition encompasses the strict interventions embraced by many nations during the pandemic, particularly those that have prevented individuals from venturing outside of their homes for most reasons.
The claims often include the idea that the benefits of lockdowns on infection control may be outweighed by the negative impacts on the economy, social structure, education and mental health. A much stronger claim that has still persistently appeared in the media as well as peer-reviewed research concerns only health effects: ... [full abstract]
Gideon Mayerowitz-Katz, Samir Bhatt, Oliver Ratmann, Jan Brauner, Seth Flaxman, Swapnil Mishra, Mrinank Sharma, Sören Mindermann, Valerie Bradley, Michaela Vollmer, Lea Merone, Gavin Yamey
BMJ Global Health, 2021, 6:e006653
[Paper]
Quantifying Ignorance in Individual-Level Causal-Effect Estimates under Hidden Confounding
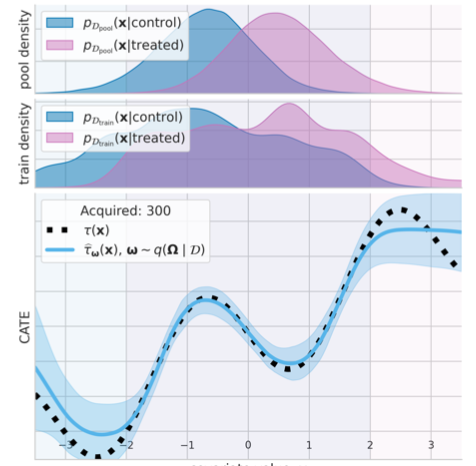
We study the problem of learning conditional average treatment effects (CATE) from high-dimensional, observational data with unobserved confounders. Unobserved confounders introduce ignorance – a level of unidentifiability – about an individual’s response to treatment by inducing bias in CATE estimates. We present a new parametric interval estimator suited for high-dimensional data, that estimates a range of possible CATE values when given a predefined bound on the level of hidden confounding. Further, previous interval estimators do not account for ignorance about the CATE stemming from samples that may be underrepresented in the original study, or samples that violate the overlap assumption. Our novel interval estimator also incorporates model uncertainty so that practitioners can be made aware of out-of-distribution data. We prove that our estimator converges to tight bounds on CATE when there may be unobserved confounding, and assess it using semi-synthetic, high-dimensional... [full abstract]
Andrew Jesson, Sören Mindermann, Yarin Gal, Uri Shalit
ICML, 2021
[arXiv]
Understanding the effectiveness of government interventions in Europe's second wave of COVID-19
As European governments face resurging waves of COVID-19, non-pharmaceutical interventions (NPIs) continue to be the primary tool for infection control. However, updated estimates of their relative effectiveness have been absent for Europe’s second wave, largely due to a lack of collated data that considers the increased subnational variation and diversity of NPIs. We collect the largest dataset of NPI implementation dates in Europe, spanning 114 subnational areas in 7 countries, with a systematic categorisation of interventions tailored to the second wave. Using a hierarchical Bayesian transmission model, we estimate the effectiveness of 17 NPIs from local case and death data. We manually validate the data, address limitations in modelling from previous studies, and extensively test the robustness of our estimates. The combined effect of all NPIs was smaller relative to estimates from the first half of 2020, indicating the strong influence of safety measures and individual prot... [full abstract]
Mrinank Sharma, Sören Mindermann, Charlie Rogers-Smith, Gavin Leech, Benedict Snodin, Janvi Ahuja, Jonas B. Sandbrink, Joshua Teperowski Monrad, George Altman, Gurpreet Dhaliwal, Lukas Finnveden, Alexander John Norman, Sebastian B. Oehm, Julia Fabienne Sandkühler, Thomas Mellan, Jan Kulveit, Leonid Chindelevitch, Seth Flaxman, Yarin Gal, Swapnil Mishra, Jan Brauner, Samir Bhatt
MedRxiv
[Paper]
Large-scale clinical interpretation of genetic variants using evolutionary data and deep learning
Quantifying the pathogenicity of protein variants in human disease-related genes would have a profound impact on clinical decisions, yet the overwhelming majority (over 98%) of these variants still have unknown consequences1–3. In principle, computational methods could support the large-scale interpretation of genetic variants. However, prior methods4–7 have relied on training machine learning models on available clinical labels. Since these labels are sparse, biased, and of variable quality, the resulting models have been considered insufficiently reliable8. By contrast, our approach leverages deep generative models to predict the clinical significance of protein variants without relying on labels. The natural distribution of protein sequences we observe across organisms is the result of billions of evolutionary experiments9,10. By modeling that distribution, we implicitly capture constraints on the protein sequences that maintain fitness. Our model EVE (Evolutionary model of V... [full abstract]
Jonathan Frazer, Pascal Notin, Mafalda Dias, Aidan Gomez, Kelly Brock, Yarin Gal, Debora S Marks
BioRXiv
[paper]
Inferring the effectiveness of government interventions against COVID-19
Governments are attempting to control the COVID-19 pandemic with nonpharmaceutical interventions (NPIs). However, the effectiveness of different NPIs at reducing transmission is poorly understood. We gathered chronological data on the implementation of NPIs for several European, and other, countries between January and the end of May 2020. We estimate the effectiveness of NPIs, ranging from limiting gathering sizes, business closures, and closure of educational institutions to stay-at-home orders. To do so, we used a Bayesian hierarchical model that links NPI implementation dates to national case and death counts and supported the results with extensive empirical validation. Closing all educational institutions, limiting gatherings to 10 people or less, and closing face-to-face businesses each reduced transmission considerably. The additional effect of stay-at-home orders was comparatively small.
Jan Brauner, Sören Mindermann, Mrinank Sharma, David Johnston, John Salvatier, Tomáš Gavenčiak, Anna B Stephenson, Gavin Leech, George Altman, Vladimir Mikulik, Alexander John Norman, Joshua Teperowski Monrad, Tamay Besiroglu, Hong Ge, Meghan A Hartwick, Yee Whye Teh, Leonid Chindelevitch, Yarin Gal, Jan Kulveit
Science (2020): eabd9338
[Paper]
Uncertainty-Aware Counterfactual Explanations for Medical Diagnosis
While deep learning algorithms can excel at predicting outcomes, they often act as black-boxes rendering them uninterpretable for healthcare practitioners. Counterfactual explanations (CEs) are a practical tool for demonstrating why machine learning models make particular decisions. We introduce a novel algorithm that leverages uncertainty to generate trustworthy counterfactual explanations for white-box models. Our proposed method can generate more interpretable CEs than the current benchmark (Van Looveren and Klaise, 2019) for breast cancer diagnosis. Further, our approach provides confidence levels for both the diagnosis as well as the explanation.
Lisa Schut, Oscar Key, Rory McGrath, Luca Costabello, Bogdan Sacaleanu, Medb Corcoran, Yarin Gal
ML4H: Machine Learning for Health Workshop NeurIPS, 2020
[Paper] [BibTex]
On the robustness of effectiveness estimation of nonpharmaceutical interventions against COVID-19 transmission
There remains much uncertainty about the relative effectiveness of different nonpharmaceutical interventions (NPIs) against COVID-19 transmission. Several studies attempt to infer NPI effectiveness with cross-country, data-driven modelling, by linking from NPI implementation dates to the observed timeline of cases and deaths in a country. These models make many assumptions. Previous work sometimes tests the sensitivity to variations in explicit epidemiological model parameters, but rarely analyses the sensitivity to the assumptions that are made by the choice the of model structure (structural sensitivity analysis). Such analysis would ensure that the inferences made are consistent under plausible alternative assumptions. Without it, NPI effectiveness estimates cannot be used to guide policy. We investigate four model structures similar to a recent state-of-the-art Bayesian hierarchical model. We find that the models differ considerably in the robustness of their NPI effectivene... [full abstract]
Mrinank Sharma, Sören Mindermann, Jan Brauner, Gavin Leech, Anna B. Stephenson, Tomáš Gavenčiak, Jan Kulveit, Yee Whye Teh, Leonid Chindelevitch, Yarin Gal
NeurIPS, 2020
[Paper]
Identifying Causal Effect Inference Failure with Uncertainty-Aware Models
Recommending the best course of action for an individual is a major application of individual-level causal effect estimation. This application is often needed in safety-critical domains such as healthcare, where estimating and communicating uncertainty to decision-makers is crucial. We introduce a practical approach for integrating uncertainty estimation into a class of state-of-the-art neural network methods used for individual-level causal estimates. We show that our methods enable us to deal gracefully with situations of “no-overlap”, common in high-dimensional data, where standard applications of causal effect approaches fail. Further, our methods allow us to handle covariate shift, where test distribution differs to train distribution, common when systems are deployed in practice. We show that when such a covariate shift occurs, correctly modeling uncertainty can keep us from giving overconfident and potentially harmful recommendations. We demonstrate our methodology with a... [full abstract]
Andrew Jesson, Sören Mindermann, Uri Shalit, Yarin Gal
NeurIPS, 2020
[arXiv] [BibTex]
Principled Uncertainty Estimation for High Dimensional Data
The ability to quantify the uncertainty in the prediction of a Bayesian deep learning model has significant practical implications—from more robust machine-learning based systems to more effective expert-in-the loop processes. While several general measures of model uncertainty exist, they are often intractable in practice when dealing with high dimensional data such as long sequences. Instead, researchers often resort to ad hoc approaches or to introducing independence assumptions to make computation tractable. We introduce a principled approach to estimate uncertainty in high dimensions that circumvents these challenges, and demonstrate its benefits in de novo molecular design.
Pascal Notin, José Miguel Hernández-Lobato, Yarin Gal
Uncertainty & Robustness in Deep Learning Workshop, ICML, 2020
[Paper]
Uncertainty Evaluation Metric for Brain Tumour Segmentation
In this paper, we develop a metric designed to assess and rank uncertainty measures for the task of brain tumour sub-tissue segmentation in the BraTS 2019 sub-challenge on uncertainty quantification. The metric is designed to: (1) reward uncertainty measures where high confidence is assigned to correct assertions, and where incorrect assertions are assigned low confidence and (2) penalize measures that have higher percentages of under-confident correct assertions. Here, the workings of the components of the metric are explored based on a number of popular uncertainty measures evaluated on the BraTS 2019 dataset.
Raghav Mehta, Angelos Filos, Yarin Gal, Tal Arbel
MIDL, 2020
[Paper]
A Systematic Comparison of Bayesian Deep Learning Robustness in Diabetic Retinopathy Tasks
Evaluation of Bayesian deep learning (BDL) methods is challenging. We often seek to evaluate the methods’ robustness and scalability, assessing whether new tools give ‘better’ uncertainty estimates than old ones. These evaluations are paramount for practitioners when choosing BDL tools on-top of which they build their applications. Current popular evaluations of BDL methods, such as the UCI experiments, are lacking: Methods that excel with these experiments often fail when used in application such as medical or automotive, suggesting a pertinent need for new benchmarks in the field. We propose a new BDL benchmark with a diverse set of tasks, inspired by a real-world medical imaging application on diabetic retinopathy diagnosis. Visual inputs (512x512 RGB images of retinas) are considered, where model uncertainty is used for medical pre-screening—i.e. to refer patients to an expert when model diagnosis is uncertain. Methods are then ranked according to metrics derived from expert... [full abstract]
Angelos Filos, Sebastian Farquhar, Aidan Gomez, Tim G. J. Rudner, Zac Kenton, Lewis Smith, Milad Alizadeh, Arnoud de Kroon, Yarin Gal
Spotlight talk, NeurIPS Workshop on Bayesian Deep Learning, 2019
[Preprint] [Code] [BibTex]

 All publications
All publications Bayesian Deep Learning
Bayesian Deep Learning Deep Learning
Deep Learning Inference
Inference Data Efficient AI
Data Efficient AI Adversarial and Interpretable ML
Adversarial and Interpretable ML Autonomous Driving
Autonomous Driving Reinforcement Learning
Reinforcement Learning Natural Language Processing
Natural Language Processing Space and Earth Observations
Space and Earth Observations Medical
Medical
 AI for Good and AI safety
AI for Good and AI safety