Back to all publications...
DiscoBAX - Discovery of optimal intervention sets in genomic experiment design
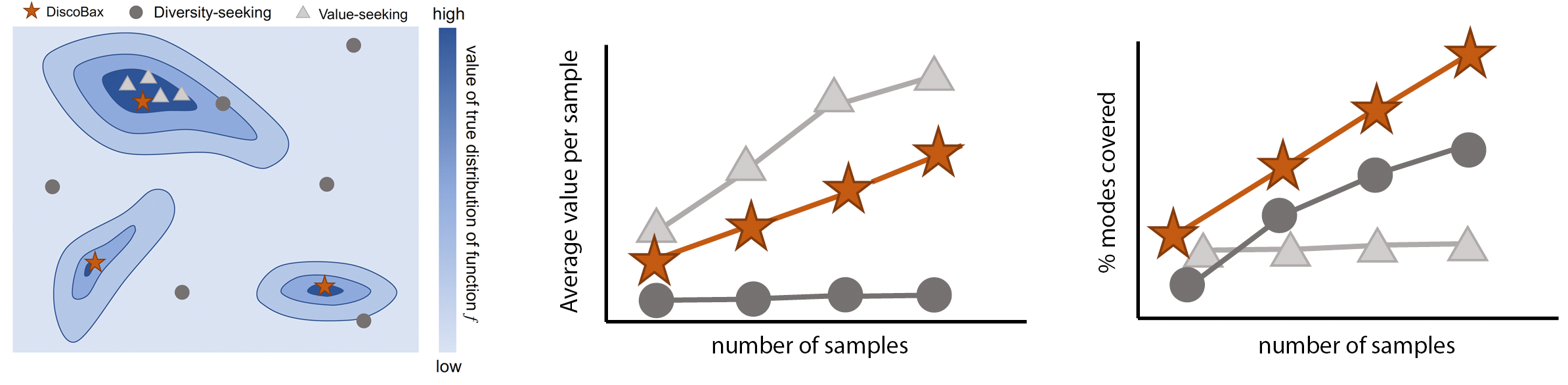
The discovery of novel therapeutics to cure genetic pathologies relies on the identification of the different genes involved in the underlying disease mechanism. With billions of potential hypotheses to test, an exhaustive exploration of the entire space of potential interventions is impossible in practice. Sample-efficient methods based on active learning or bayesian optimization bear the promise of identifying interesting targets using the least experiments possible. However, genomic perturbation experiments typically rely on proxy outcomes measured in biological model systems that may not completely correlate with the outcome of interventions in humans. In practical experiment design, one aims to find a set of interventions which maximally move a target phenotype via a diverse set of mechanisms in order to reduce the risk of failure in future stages of trials. To that end, we introduce DiscoBAX — a sample-efficient algorithm for the discovery of genetic interventions that maximize the movement of a phenotype in a direction of interest while covering a diverse set of underlying mechanisms. We provide theoretical guarantees on the optimality of the approach under standard assumptions, conduct extensive experiments in synthetic and real-world settings relevant to genomic discovery and demonstrate that DiscoBAX outperforms state-of-the-art active learning and Bayesian optimization methods in this task. Better methods for selecting effective and diverse perturbations in biological systems could enable researchers to potentially discover novel therapeutics for a range of genetically-driven diseases.
Clare Lyle, Arash Mehrjou, Pascal Notin, Andrew Jesson, Stefan Bauer, Yarin Gal, Patrick Schwab
ICML 2023
[arXiv]