Back to all publications...
ProteinGym: Large-Scale Benchmarks for Protein Fitness Prediction and Design
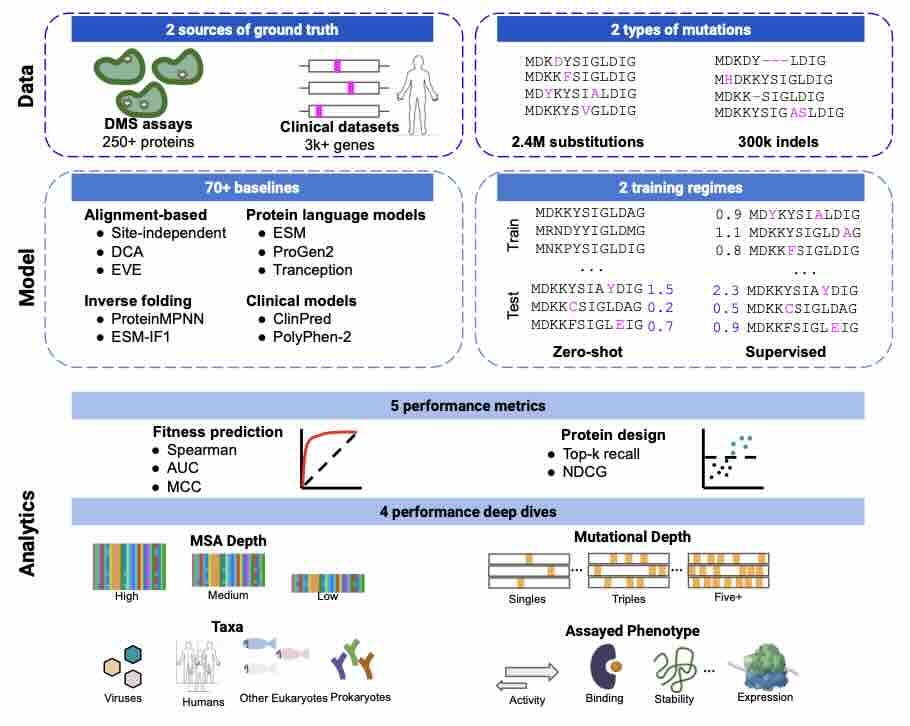
Predicting the effects of mutations in proteins is critical to many applications, from understanding genetic disease to designing novel proteins that can address our most pressing challenges in climate, agriculture and healthcare. Despite a surge in machine learning-based protein models to tackle these questions, an assessment of their respective benefits is challenging due to the use of distinct, often contrived, experimental datasets, and the variable performance of models across different protein families. Addressing these challenges requires scale. To that end we introduce ProteinGym, a large-scale and holistic set of benchmarks specifically designed for protein fitness prediction and design. It encompasses both a broad collection of over 250 standardized deep mutational scanning assays, spanning millions of mutated sequences, as well as curated clinical datasets providing high- quality expert annotations about mutation effects. We devise a robust evaluation framework that combines metrics for both fitness prediction and design, factors in known limitations of the underlying experimental methods, and covers both zero-shot and supervised settings. We report the performance of a diverse set of over 70 high-performing models from various subfields (eg., alignment-based, inverse folding) into a unified benchmark suite. We open source the corresponding codebase, datasets, MSAs, structures, model predictions and develop a user-friendly website that facilitates data access and analysis.
Pascal Notin, Aaron W. Kollasch, Daniel Ritter, Lood van Niekerk, Steffanie Paul, Hansen Spinner, Nathan Rollins, Ada Shaw, Ruben Weitzman, Jonathan Frazer, Mafalda Dias, Dinko Franceschi, Rose Orenbuch, Yarin Gal, Debora Marks
NeurIPS 2023
[Paper]