Back to all publications...
ProteinNPT: Improving Protein Property Prediction and Design with Non-Parametric Transformers
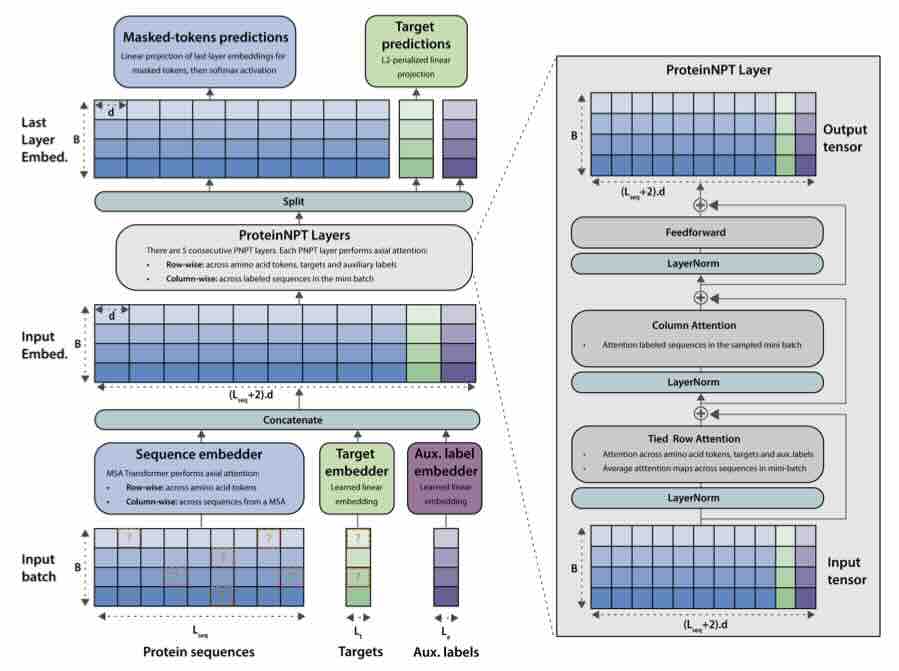
Protein design holds immense potential for optimizing naturally occurring proteins, with broad applications in drug discovery, material design, and sustainability. How- ever, computational methods for protein engineering are confronted with significant challenges, such as an expansive design space, sparse functional regions, and a scarcity of available labels. These issues are further exacerbated in practice by the fact most real-life design scenarios necessitate the simultaneous optimization of multiple properties. In this work, we introduce ProteinNPT, a non-parametric trans- former variant tailored to protein sequences and particularly suited to label-scarce and multi-task learning settings. We first focus on the supervised fitness prediction setting and develop several cross-validation schemes which support robust perfor- mance assessment. We subsequently reimplement prior top-performing baselines, introduce several extensions of these baselines by integrating diverse branches of the protein engineering literature, and demonstrate that ProteinNPT consistently outperforms all of them across a diverse set of protein property prediction tasks. Finally, we demonstrate the value of our approach for iterative protein design across extensive in silico Bayesian optimization and conditional sampling experiments.
Pascal Notin, Ruben Weitzman, Debora Marks, Yarin Gal
NeurIPS 2023
[Paper]