Back to all members...
Ruben Weitzman
PhD, started 2021

Ruben is a DPhil candidate with the Health Data Science CDT, and is co-supervised by Yarin Gal and Debora Marks. He is interested in studying gene regulation at different scales and multi-disciplinary projects at the interface between machine learning and bioinformatics. He is funded by an EPSRC and Kellogg Scholarship. Ruben previously completed an MPhil in Computational Biology at the University of Cambridge, and a BE in Bioengineering and Biomedical Engineering at Imperial College London.
Publications while at OATML • News items mentioning Ruben Weitzman • Reproducibility and Code • Blog Posts
Publications while at OATML:
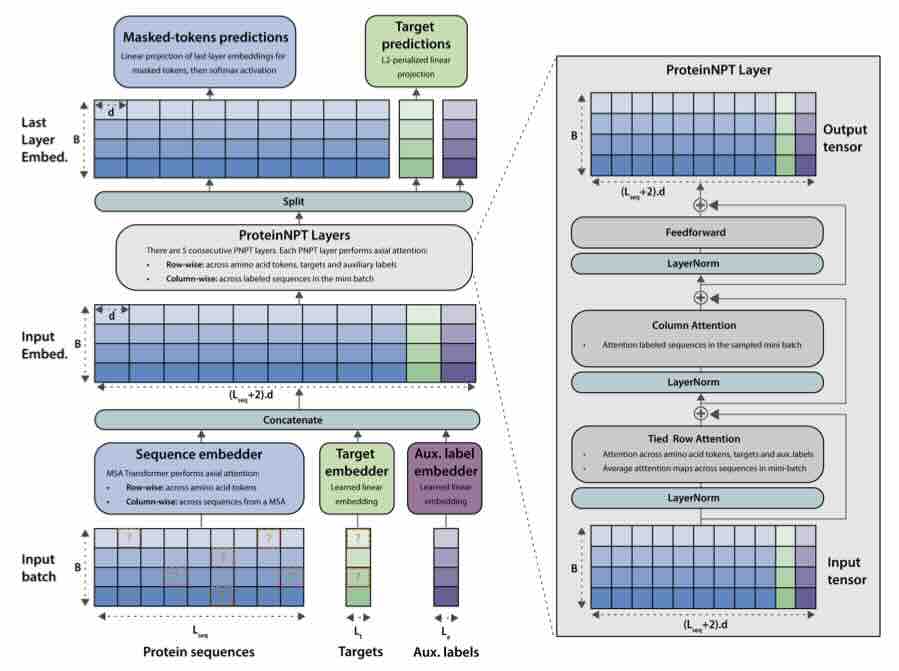
ProteinNPT: Improving Protein Property Prediction and Design with Non-Parametric Transformers
Protein design holds immense potential for optimizing naturally occurring proteins, with broad applications in drug discovery, material design, and sustainability. How- ever, computational methods for protein engineering are confronted with significant challenges, such as an expansive design space, sparse functional regions, and a scarcity of available labels. These issues are further exacerbated in practice by the fact most real-life design scenarios necessitate the simultaneous optimization of multiple properties. In this work, we introduce ProteinNPT, a non-parametric trans- former variant tailored to protein sequences and particularly suited to label-scarce and multi-task learning settings. We first focus on the supervised fitness prediction setting and develop several cross-validation schemes which support robust perfor- mance assessment. We subsequently reimplement prior top-performing baselines, introduce several extensions of these baselines by integrating diverse branches ... [full abstract]
Pascal Notin, Ruben Weitzman, Debora Marks, Yarin Gal
NeurIPS 2023
[Paper]
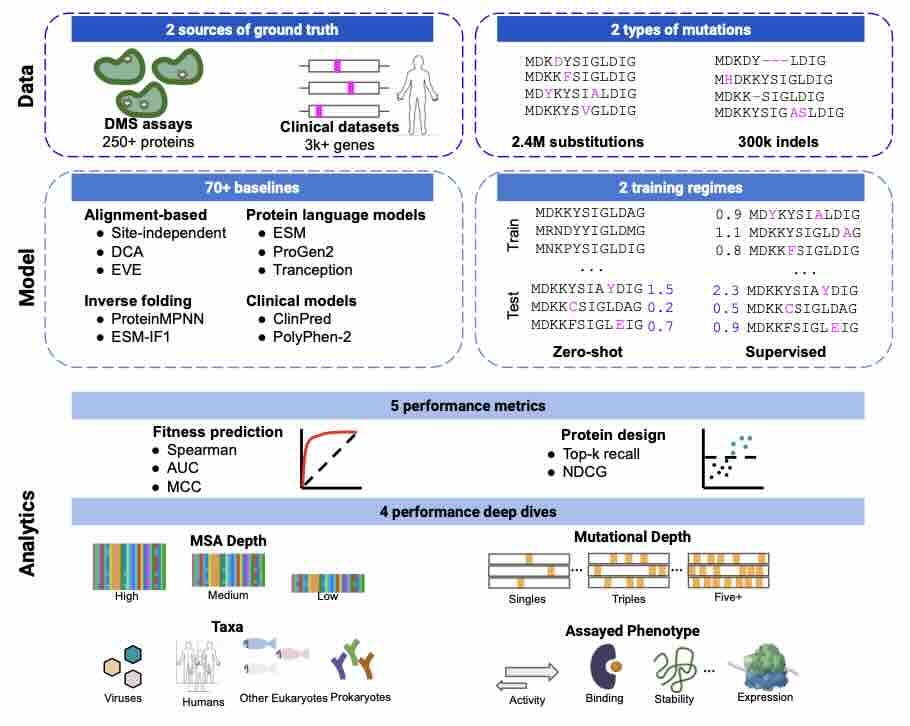
ProteinGym: Large-Scale Benchmarks for Protein Fitness Prediction and Design
Predicting the effects of mutations in proteins is critical to many applications, from understanding genetic disease to designing novel proteins that can address our most pressing challenges in climate, agriculture and healthcare. Despite a surge in machine learning-based protein models to tackle these questions, an assessment of their respective benefits is challenging due to the use of distinct, often contrived, experimental datasets, and the variable performance of models across different protein families. Addressing these challenges requires scale. To that end we introduce ProteinGym, a large-scale and holistic set of benchmarks specifically designed for protein fitness prediction and design. It encompasses both a broad collection of over 250 standardized deep mutational scanning assays, spanning millions of mutated sequences, as well as curated clinical datasets providing high- quality expert annotations about mutation effects. We devise a robust evaluation framework that comb... [full abstract]
Pascal Notin, Aaron W. Kollasch, Daniel Ritter, Lood van Niekerk, Steffanie Paul, Hansen Spinner, Nathan Rollins, Ada Shaw, Ruben Weitzman, Jonathan Frazer, Mafalda Dias, Dinko Franceschi, Rose Orenbuch, Yarin Gal, Debora Marks
NeurIPS 2023
[Paper]
News items mentioning Ruben Weitzman:

OATML to co-organize the Workshop on Computational Biology (WCB) at ICML 2023
30 Mar 2023
OATML students Pascal Notin and Ruben Wietzman are co-organizing the Workshop on Computational Biology (WCB) at ICML 2023 jointly with collaborators at Harvard, Columbia, Cornell and others. OATML students Freddie Bickford Smith, Jan Brauner and Shreshth Malik are part of the program committee.

OATML to co-organize the Machine Learning for Drug Discovery (MLDD) workshop at ICLR 2023
21 Dec 2022
OATML students Pascal Notin and Clare Lyle, along with OATML group leader Yarin Gal, are co-organizing the Machine Learning for Drug Discovery (MLDD) workshop at ICLR 2023 jointly with collaborators at GSK, Genentech, Harvard, MIT and others. OATML students Neil Band, Freddie Bickford Smith, Jan Brauner, Lars Holdijk, Andrew Jesson, Andreas Kirsch, Shreshth Malik, Lood van Niekirk and Ruben Wietzman are part of the program committee.