Back to all members...
Pascal Notin
Associate Member (Senior Research Fellow), started 2019

Pascal is a Scientific Lead in the Marks lab at Harvard Medical school, where he leads the unit focusing on protein engineering. He is passionate about the use of machine learning models to design novel biomolecules to address challenges in healthcare and sustainability.
His research lies at the intersection of generative AI, computational biology and chemistry. He was a PhD student in the Oxford Applied and Theoretical Machine Learning Group, part of the CS Department at the University of Oxford, under the supervision of Yarin Gal. During his time at Oxford, he developed large-scale protein language models for fitness prediction and design (1 2 3 4), and deep generative models to predict the impact of genetic mutations in humans (5) or viral mutations likely to escape immunity (6).
Before Oxford, he was a Senior Manager at McKinsey & Company in New York and Paris, where he was leading cross-disciplinary teams on high-impact analytics projects. He holds an M.S. in Operations Research from Columbia University and a B.S. and M.S. in Applied Mathematics and Physics from Ecole Polytechnique. Combining rigorous academic training with practical industry experience, Pascal is now focused on driving innovations in bioengineering, resolutely committed to creating a healthier and more sustainable future.
Publications while at OATML • News items mentioning Pascal Notin • Reproducibility and Code • Blog Posts
Publications while at OATML:
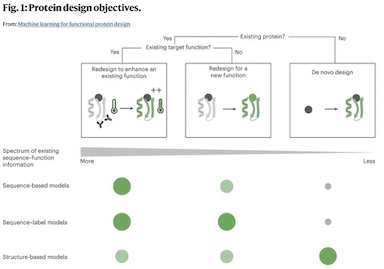
Machine learning for functional protein design
Recent breakthroughs in AI coupled with the rapid accumulation of protein sequence and structure data have radically transformed computational protein design. New methods promise to escape the constraints of natural and laboratory evolution, accelerating the generation of proteins for applications in biotechnology and medicine. To make sense of the exploding diversity of machine learning approaches, we introduce a unifying framework that classifies models on the basis of their use of three core data modalities: sequences, structures and functional labels. We discuss the new capabilities and outstanding challenges for the practical design of enzymes, antibodies, vaccines, nanomachines and more. We then highlight trends shaping the future of this field, from large-scale assays to more robust benchmarks, multimodal foundation models, enhanced sampling strategies and laboratory automation.
Pascal Notin, Nathan Rollins, Yarin Gal, Chris Sander, Debora Marks
Nature Biotechnology (2024)
[paper]
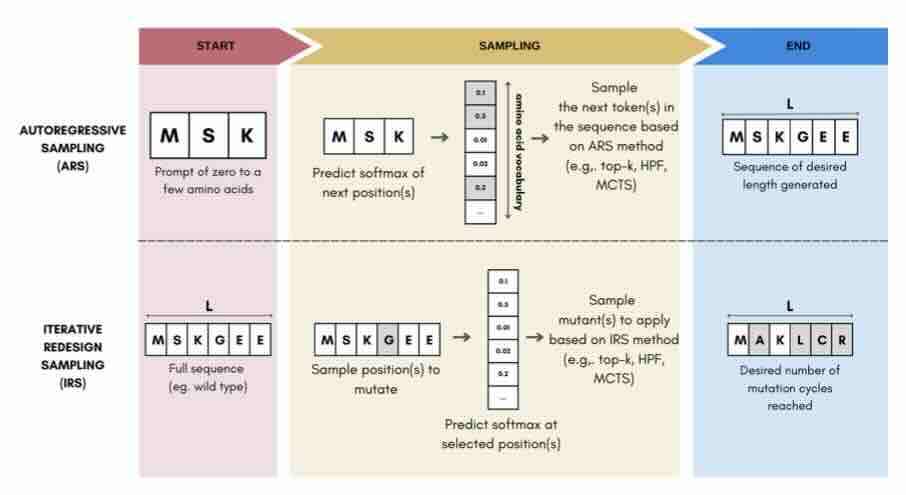
Sampling Protein Language Models for Functional Protein Design
Protein language models have emerged as powerful ways to learn complex repre- sentations of proteins, thereby improving their performance on several downstream tasks, from structure prediction to fitness prediction, property prediction, homology detection, and more. By learning a distribution over protein sequences, they are also very promising tools for designing novel and functional proteins, with broad applications in healthcare, new material, or sustainability. Given the vastness of the corresponding sample space, efficient exploration methods are critical to the success of protein engineering efforts. However, the methodologies for ade- quately sampling these models to achieve core protein design objectives remain underexplored and have predominantly leaned on techniques developed for Natural Language Processing. In this work, we first develop a holistic in silico protein design evaluation framework, to comprehensively compare different sampling methods. After performing a tho... [full abstract]
Jeremie Theddy Darmawan, Yarin Gal, Pascal Notin
Machine Learning for Structural Biology / Generative AI and Biology workshops, NeurIPS 2023
[Paper]
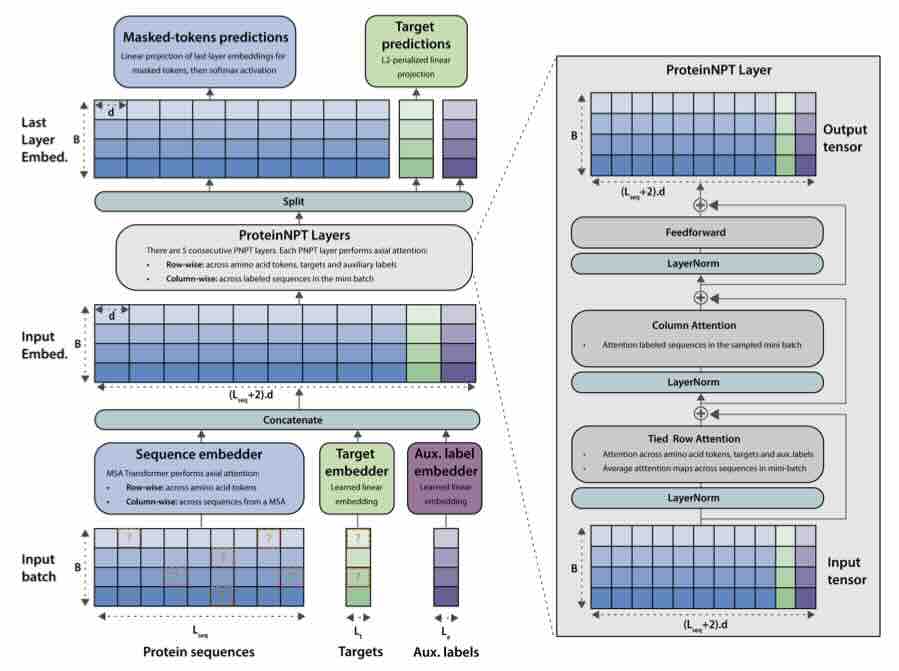
ProteinNPT: Improving Protein Property Prediction and Design with Non-Parametric Transformers
Protein design holds immense potential for optimizing naturally occurring proteins, with broad applications in drug discovery, material design, and sustainability. How- ever, computational methods for protein engineering are confronted with significant challenges, such as an expansive design space, sparse functional regions, and a scarcity of available labels. These issues are further exacerbated in practice by the fact most real-life design scenarios necessitate the simultaneous optimization of multiple properties. In this work, we introduce ProteinNPT, a non-parametric trans- former variant tailored to protein sequences and particularly suited to label-scarce and multi-task learning settings. We first focus on the supervised fitness prediction setting and develop several cross-validation schemes which support robust perfor- mance assessment. We subsequently reimplement prior top-performing baselines, introduce several extensions of these baselines by integrating diverse branches ... [full abstract]
Pascal Notin, Ruben Weitzman, Debora Marks, Yarin Gal
NeurIPS 2023
[Paper]
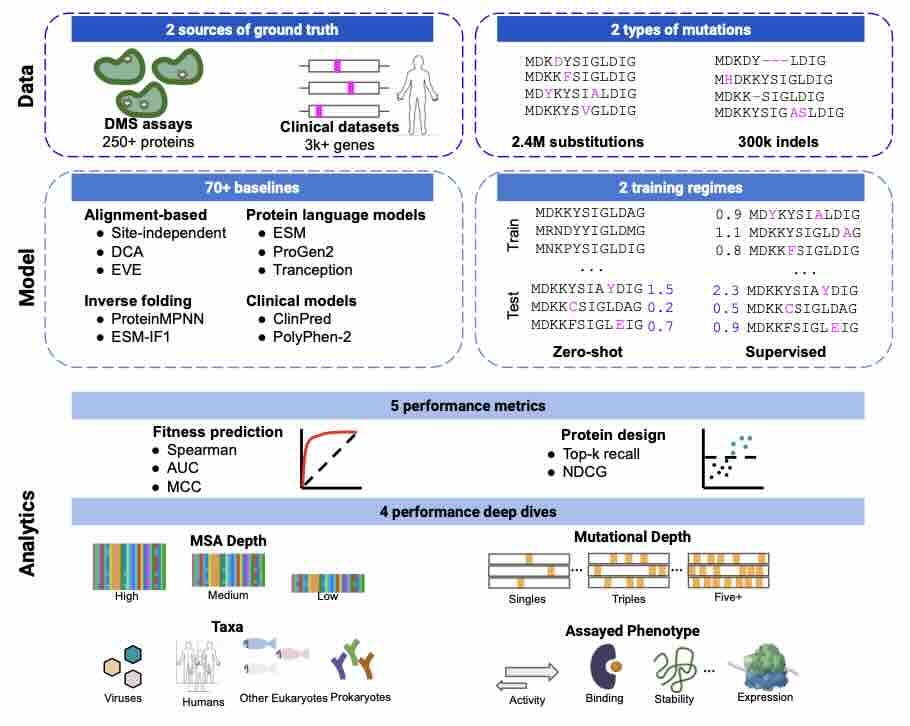
ProteinGym: Large-Scale Benchmarks for Protein Fitness Prediction and Design
Predicting the effects of mutations in proteins is critical to many applications, from understanding genetic disease to designing novel proteins that can address our most pressing challenges in climate, agriculture and healthcare. Despite a surge in machine learning-based protein models to tackle these questions, an assessment of their respective benefits is challenging due to the use of distinct, often contrived, experimental datasets, and the variable performance of models across different protein families. Addressing these challenges requires scale. To that end we introduce ProteinGym, a large-scale and holistic set of benchmarks specifically designed for protein fitness prediction and design. It encompasses both a broad collection of over 250 standardized deep mutational scanning assays, spanning millions of mutated sequences, as well as curated clinical datasets providing high- quality expert annotations about mutation effects. We devise a robust evaluation framework that comb... [full abstract]
Pascal Notin, Aaron W. Kollasch, Daniel Ritter, Lood van Niekerk, Steffanie Paul, Hansen Spinner, Nathan Rollins, Ada Shaw, Ruben Weitzman, Jonathan Frazer, Mafalda Dias, Dinko Franceschi, Rose Orenbuch, Yarin Gal, Debora Marks
NeurIPS 2023
[Paper]
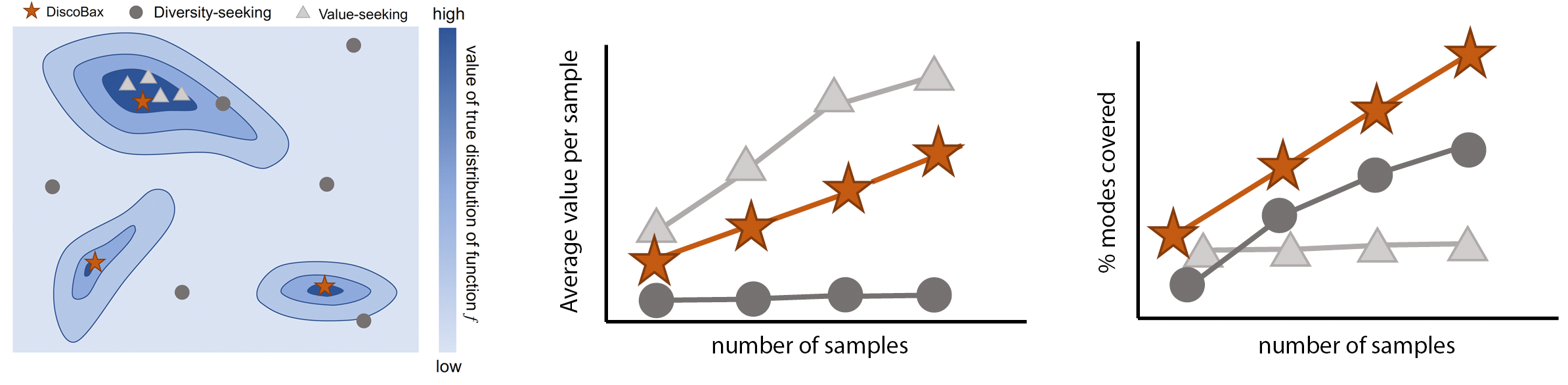
DiscoBAX - Discovery of optimal intervention sets in genomic experiment design
The discovery of novel therapeutics to cure genetic pathologies relies on the identification of the different genes involved in the underlying disease mechanism. With billions of potential hypotheses to test, an exhaustive exploration of the entire space of potential interventions is impossible in practice. Sample-efficient methods based on active learning or bayesian optimization bear the promise of identifying interesting targets using the least experiments possible. However, genomic perturbation experiments typically rely on proxy outcomes measured in biological model systems that may not completely correlate with the outcome of interventions in humans. In practical experiment design, one aims to find a set of interventions which maximally move a target phenotype via a diverse set of mechanisms in order to reduce the risk of failure in future stages of trials. To that end, we introduce DiscoBAX — a sample-efficient algorithm for the discovery of genetic interventions that maxim... [full abstract]
Clare Lyle, Arash Mehrjou, Pascal Notin, Andrew Jesson, Stefan Bauer, Yarin Gal, Patrick Schwab
ICML 2023
[arXiv]
Mixtures of large-scale dynamic functional brain network modes
Accurate temporal modelling of functional brain networks is essential in the quest for understanding how such networks facilitate cognition. Researchers are beginning to adopt time-varying analyses for electrophysiological data that capture highly dynamic processes on the order of milliseconds. Typically, these approaches, such as clustering of functional connectivity profiles and Hidden Markov Modelling (HMM), assume mutual exclusivity of networks over time. Whilst a powerful constraint, this assumption may be compromising the ability of these approaches to describe the data effectively. Here, we propose a new generative model for functional connectivity as a time-varying linear mixture of spatially distributed statistical "modes". The temporal evolution of this mixture is governed by a recurrent neural network, which enables the model to generate data with a rich temporal structure. We use a Bayesian framework known as amortised variational inference to learn model parameters fro... [full abstract]
Chetan Gohil, Evan Roberts, Ryan Timms, Alex Skates, Cameron Higgins, Andrew Quinn, Usama Pervaiz, Joost van Amersfoort, Pascal Notin, Yarin Gal, Stanislaw Adaszewski, Mark Woolrich
NeuroImage
[paper]
Learning from pre-pandemic data to forecast viral antibody escape
From early detection of variants of concern to vaccine and therapeutic design, pandemic preparedness depends on identifying viral mutations that escape the response of the host immune system. While experimental scans are useful for quantifying escape potential, they remain laborious and impractical for exploring the combinatorial space of mutations. Here we introduce a biologically grounded model to quantify the viral escape potential of mutations at scale. Our method - EVEscape - brings together fitness predictions from evolutionary models, structure-based features that assess antibody binding potential, and distances between mutated and wild-type residues. Unlike other models that predict variants of concern based on newly observed variants, EVEscape has no reliance on recent community prevalence, and is applicable before surveillance sequencing or experimental scans are broadly available. We validate EVEscape predictions against experimental data on H1N1, HIV and SARS-CoV-2, inc... [full abstract]
Nicole N Thadani, Sarah Gurev, Pascal Notin, Noor Youssef, Nathan J Rollins, Daniel Ritter, Chris Sander, Yarin Gal, Debora Marks
Nature
[paper]
Tranception: protein fitness prediction with autoregressive transformers and inference-time retrieval
The ability to accurately model the fitness landscape of protein sequences is critical to a wide range of applications, from quantifying the effects of human variants on disease likelihood, to predicting immune-escape mutations in viruses and designing novel biotherapeutic proteins. Deep generative models of protein sequences trained on multiple sequence alignments have been the most successful approaches so far to address these tasks. The performance of these methods is however contingent on the availability of sufficiently deep and diverse alignments for reliable training. Their potential scope is thus limited by the fact many protein families are hard, if not impossible, to align. Large language models trained on massive quantities of non-aligned protein sequences from diverse families address these problems and show potential to eventually bridge the performance gap. We introduce Tranception, a novel transformer architecture leveraging autoregressive predictions and retrieval o... [full abstract]
Pascal Notin, Mafalda Dias, Jonathan Frazer, Javier Marchena-Hurtado, Aidan Gomez, Debora Marks, Yarin Gal
ICML, 2022
[Preprint] [BibTex] [Code]
RITA: a Study on Scaling Up Generative Protein Sequence Models
In this work we introduce RITA: a suite of autoregressive generative models for protein sequences, with up to 1.2 billion parameters, trained on over 280 million protein sequences belonging to the UniRef-100 database. Such generative models hold the promise of greatly accelerating protein design. We conduct the first systematic study of how capabilities evolve with model size for autoregressive transformers in the protein domain: we evaluate RITA models in next amino acid prediction, zero-shot fitness, and enzyme function prediction, showing benefits from increased scale. We release the RITA models openly, to the benefit of the research community.
Daniel Hesslow, Niccoló Zanichelli, Pascal Notin, Iacopo Poli, Debora Marks
ICML, Workshop on Computational biology, 2022 (Spotlight)
[Preprint] [Code] [BibTex]
GeneDisco: A Benchmark for Experimental Design in Drug Discovery
In vitro cellular experimentation with genetic interventions, using for example CRISPR technologies, is an essential step in early-stage drug discovery and target validation that serves to assess initial hypotheses about causal associations between biological mechanisms and disease pathologies. With billions of potential hypotheses to test, the experimental design space for in vitro genetic experiments is extremely vast, and the available experimental capacity - even at the largest research institutions in the world - pales in relation to the size of this biological hypothesis space. Machine learning methods, such as active and reinforcement learning, could aid in optimally exploring the vast biological space by integrating prior knowledge from various information sources as well as extrapolating to yet unexplored areas of the experimental design space based on available data. However, there exist no standardised benchmarks and data sets for this challenging task and little researc... [full abstract]
Arash Mehrjou, Ashkan Soleymani, Andrew Jesson, Pascal Notin, Yarin Gal, Stefan Bauer, Patrick Schwab
International Conference on Learning Representations, 2022
[Preprint] [BibTex] [Code]
Mixtures of large-scale dynamic functional brain network modes
Accurate temporal modelling of functional brain networks is essential in the quest for understanding how such networks facilitate cognition. Researchers are beginning to adopt time-varying analyses for electrophysiological data that capture highly dynamic processes on the order of milliseconds. Typically, these approaches, such as clustering of functional connectivity profiles and Hidden Markov Modelling (HMM), assume mutual exclusivity of networks over time. Whilst a powerful constraint, this assumption may be compromising the ability of these approaches to describe the data effectively. Here, we propose a new generative model for functional connectivity as a time-varying linear mixture of spatially distributed statistical “modes”. The temporal evolution of this mixture is governed by a recurrent neural network, which enables the model to generate data with a rich temporal structure. We use a Bayesian framework known as amortised variational inference to learn model parameters fro... [full abstract]
Chetan Gohil, Evan Roberts, Ryan Timms, Alex Skates, Cameron Higgins, Andrew Quinn, Usama Pervaiz, Joost van Amersfoort, Pascal Notin, Yarin Gal, Stanislaw Adaszewski, Mark Woolrich
NeuroImage Volume 263 [Paper] [BibTex]
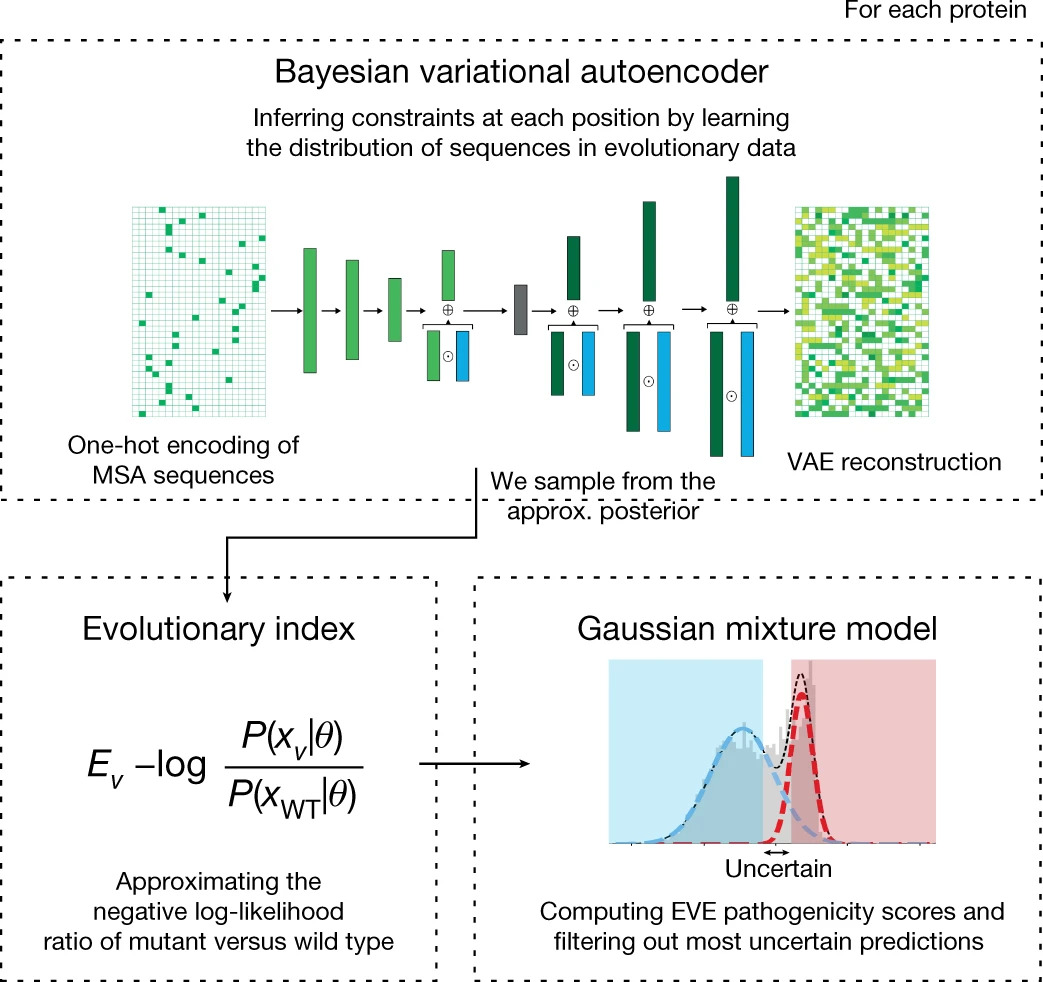
Disease variant prediction with deep generative models of evolutionary data
Quantifying the pathogenicity of protein variants in human disease-related genes would have a marked effect on clinical decisions, yet the overwhelming majority (over 98%) of these variants still have unknown consequences. In principle, computational methods could support the large-scale interpretation of genetic variants. However, state-of-the-art methods have relied on training machine learning models on known disease labels. As these labels are sparse, biased and of variable quality, the resulting models have been considered insufficiently reliable. Here we propose an approach that leverages deep generative models to predict variant pathogenicity without relying on labels. By modelling the distribution of sequence variation across organisms, we implicitly capture constraints on the protein sequences that maintain fitness. Our model EVE (evolutionary model of variant effect) not only outperforms computational approaches that rely on labelled data but also performs on par with, if... [full abstract]
Jonathan Frazer, Pascal Notin, Mafalda Dias, Aidan Gomez, Joseph K. Min, Kelly Brock, Yarin Gal, Debora Marks
Nature, 2021 (volume 599, pages 91–95)
[Paper] [BibTex] [Preprint] [Website] [Code]
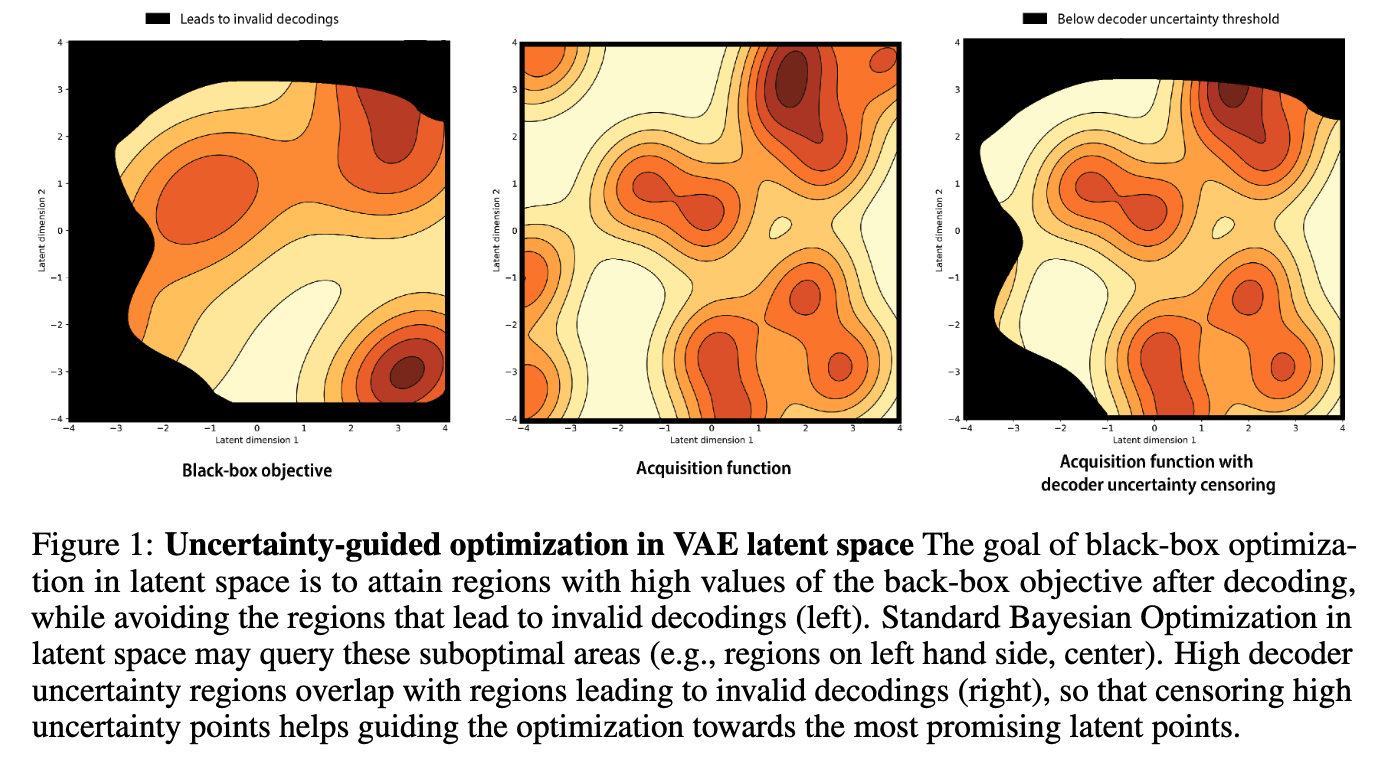
Improving black-box optimization in VAE latent space using decoder uncertainty
Optimization in the latent space of variational autoencoders is a promising approach to generate high-dimensional discrete objects that maximize an expensive black-box property (e.g., drug-likeness in molecular generation, function approximation with arithmetic expressions). However, existing methods lack robustness as they may decide to explore areas of the latent space for which no data was available during training and where the decoder can be unreliable, leading to the generation of unrealistic or invalid objects. We propose to leverage the epistemic uncertainty of the decoder to guide the optimization process. This is not trivial though, as a naive estimation of uncertainty in the high-dimensional and structured settings we consider would result in high estimator variance. To solve this problem, we introduce an importance sampling-based estimator that provides more robust estimates of epistemic uncertainty. Our uncertainty-guided optimization approach does not require modifica... [full abstract]
Pascal Notin, José Miguel Hernández-Lobato, Yarin Gal
NeurIPS, 2021
[Preprint] [Proceedings] [BibTex] [Code]
Large-scale clinical interpretation of genetic variants using evolutionary data and deep learning
Quantifying the pathogenicity of protein variants in human disease-related genes would have a profound impact on clinical decisions, yet the overwhelming majority (over 98%) of these variants still have unknown consequences1–3. In principle, computational methods could support the large-scale interpretation of genetic variants. However, prior methods4–7 have relied on training machine learning models on available clinical labels. Since these labels are sparse, biased, and of variable quality, the resulting models have been considered insufficiently reliable8. By contrast, our approach leverages deep generative models to predict the clinical significance of protein variants without relying on labels. The natural distribution of protein sequences we observe across organisms is the result of billions of evolutionary experiments9,10. By modeling that distribution, we implicitly capture constraints on the protein sequences that maintain fitness. Our model EVE (Evolutionary model of Vari... [full abstract]
Jonathan Frazer, Pascal Notin, Mafalda Dias, Aidan Gomez, Kelly Brock, Yarin Gal, Debora S Marks
BioRXiv
[paper]
Principled Uncertainty Estimation for High Dimensional Data
The ability to quantify the uncertainty in the prediction of a Bayesian deep learning model has significant practical implications—from more robust machine-learning based systems to more effective expert-in-the loop processes. While several general measures of model uncertainty exist, they are often intractable in practice when dealing with high dimensional data such as long sequences. Instead, researchers often resort to ad hoc approaches or to introducing independence assumptions to make computation tractable. We introduce a principled approach to estimate uncertainty in high dimensions that circumvents these challenges, and demonstrate its benefits in de novo molecular design.
Pascal Notin, José Miguel Hernández-Lobato, Yarin Gal
Uncertainty & Robustness in Deep Learning Workshop, ICML, 2020
[Paper]
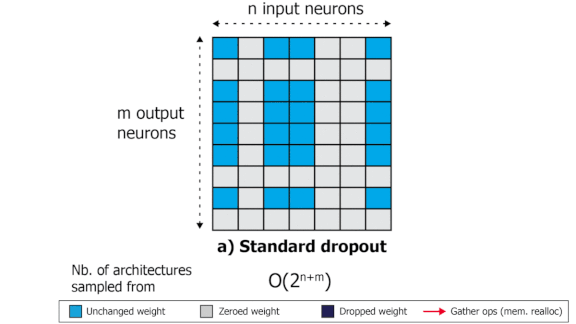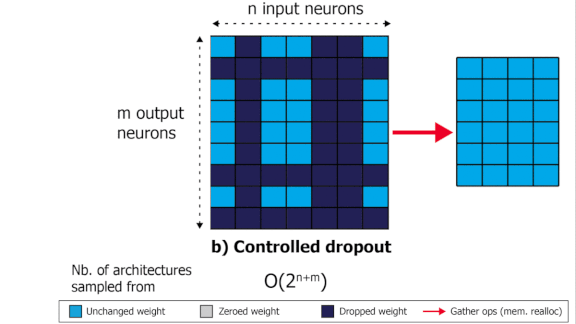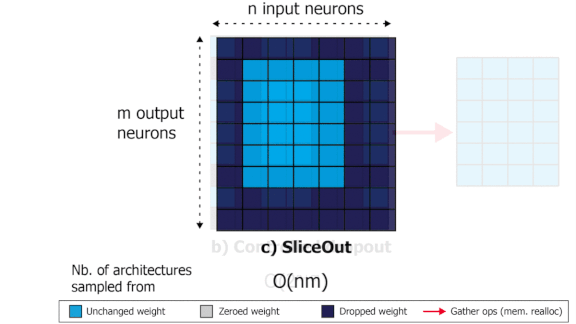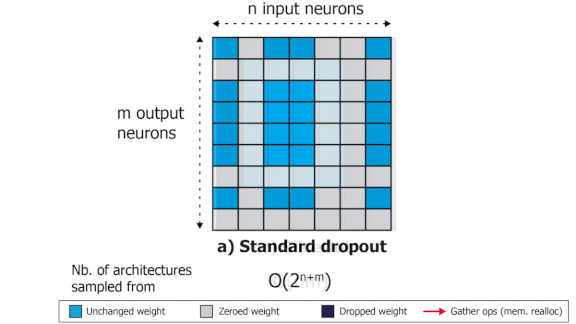
SliceOut: Training Transformers and CNNs faster while using less memory
We demonstrate 10-40% speedups and memory reduction with Wide ResNets, EfficientNets, and Transformer models, with minimal to no loss in accuracy, using SliceOut---a new dropout scheme designed to take advantage of GPU memory layout. By dropping contiguous sets of units at random, our method preserves the regularization properties of dropout while allowing for more efficient low-level implementation, resulting in training speedups through (1) fast memory access and matrix multiplication of smaller tensors, and (2) memory savings by avoiding allocating memory to zero units in weight gradients and activations. Despite its simplicity, our method is highly effective. We demonstrate its efficacy at scale with Wide ResNets & EfficientNets on CIFAR10/100 and ImageNet, as well as Transformers on the LM1B dataset. These speedups and memory savings in training can lead to CO2 emissions reduction of up to 40% for training large models.
Pascal Notin, Aidan Gomez, Joanna Yoo, Yarin Gal
Under review
[Paper]
News items mentioning Pascal Notin:

OATML to co-organize the Workshop on Computational Biology (WCB) at ICML 2023
30 Mar 2023
OATML students Pascal Notin and Ruben Wietzman are co-organizing the Workshop on Computational Biology (WCB) at ICML 2023 jointly with collaborators at Harvard, Columbia, Cornell and others. OATML students Freddie Bickford Smith, Jan Brauner and Shreshth Malik are part of the program committee.

OATML to co-organize the Machine Learning for Drug Discovery (MLDD) workshop at ICLR 2023
21 Dec 2022
OATML students Pascal Notin and Clare Lyle, along with OATML group leader Yarin Gal, are co-organizing the Machine Learning for Drug Discovery (MLDD) workshop at ICLR 2023 jointly with collaborators at GSK, Genentech, Harvard, MIT and others. OATML students Neil Band, Freddie Bickford Smith, Jan Brauner, Lars Holdijk, Andrew Jesson, Andreas Kirsch, Shreshth Malik, Lood van Niekirk and Ruben Wietzman are part of the program committee.

Pascal Notin to present at the ELLIS unit in Copenhagen
17 Jun 2022
OATML DPhil student Pascal Notin will give a talk at the Center for Basic Machine Learning Research in Life Science (MLLS), ELLIS Unit in Copenhagen, on generative models for protein fitness and mutation effects prediction.

OATML to co-organize the Workshop on Computational Biology at ICML 2022
04 May 2022
OATML student Pascal Notin is co-organizing the 7th edition of the Workshop on Computational Biology (WCB) at ICML 2022 jointly with collaborators at Harvard, Columbia, Cornell and others. OATML students Neil Band, Freddie Bickford Smith, Jan Brauner, Andreas Kirsch and Lood van Niekirk are part of the PC.


OATML to co-organize the Machine Learning for Drug Discovery (MLDD) workshop at ICLR 2022
15 Jan 2022
OATML students Pascal Notin, Andrew Jesson and Clare Lyle, along with OATML group leader Professor Yarin Gal, are co-organizing the first Machine Learning for Drug Discovery (MLDD) workshop at ICLR 2022 jointly with collaborators at GSK, Harvard, MILA, MIT and others. OATML students Neil Band, Freddie Bickford Smith, Jan Brauner, Lars Holdijk, Andreas Kirsch, Jannik Kossen and Muhammed Razzak are part of the PC.

OATML researchers publish paper in Nature
03 Nov 2021
The paper “Disease variant prediction with deep generative models of evolutionary data” was published in Nature. The work is a collaboration between OATML and the Marks lab at Harvard Medical School. It was led by Pascal Notin and Professor Yarin Gal from OATML together with Jonathan Frazer, Mafalda Dias, and Debbie Marks from the Marks lab. OATML DPhil student Aidan Gomez and Marks lab researchers Joseph Min and Kelly Brock are co-authors on the paper.

NeurIPS 2021
11 Oct 2021
Thirteen papers with OATML members accepted to NeurIPS 2021 main conference. More information in our blog post.

OATML graduate students receive best reviewer awards and serve as expert reviewers at ICML 2021
06 Sep 2021
OATML graduate students Sebastian Farquhar and Jannik Kossen receive best reviewer awards (top 10%) at ICML 2021. Further, OATML graduate students Tim G. J. Rudner, Pascal Notin, Panagiotis Tigas, and Binxin Ru have served the conference as expert reviewers.

OATML student receives a best poster award at the ICML workshop on Computational Biology
07 Aug 2021
OATML MSc student Lood van Niekerk received a best poster award at the 2021 ICML Workshop on Computational Biology for his work on “Exploring the latent space of deep generative models: Applications to G-protein coupled receptors”. This is part of an ongoing collaboration between OATML and the Marks Lab. From OATML, Professor Yarin Gal and DPhil student Pascal Notin are co-authors on the paper.

ICML 2021
17 Jul 2021
Seven papers with OATML members accepted to ICML 2021, together with 14 workshop papers. More information in our blog post.

OATML graduate students receive Outstanding Reviewer Awards
03 Jun 2021
OATML graduate students Pascal Notin and Tim G. J. Rudner received best reviewer awards (top 5%) at UAI 2021.

OATML student invited to speak at the Cornell ML in Medicine seminar series
19 Mar 2021
OATML graduate student Pascal Notin will give an invited talk on Uncertainty in deep generative models with applications to genomics and drug design at the Cornell Machine Learning in Medicine seminar series. Professor Yarin Gal and collaborator José Miguel Hernández-Lobato are co-authors on the paper.

OATML student invited to speak at the EMBL workshop on ML in Drug Discovery
11 Mar 2021
OATML graduate student Pascal Notin will give an invited talk on “Large-scale clinical interpretation of genetic variants using evolutionary data and deep generative models” at the EMBL-EBI workshop on Machine Learning in Drug Discovery. The work is a collaboration between OATML (Pascal and Professor Yarin Gal) and the Marks Lab at Harvard.
Reproducibility and Code
Tranception: protein fitness prediction with autoregressive transformers and inference-time retrieval
This is the official code repository for the paper “Tranception: protein fitness prediction with autoregressive transformers and inference-time retrieval”. The repository also includes the full ProteinGym benchmarks.
CodePascal Notin
Improving black-box optimization in VAE latent space using decoder uncertainty
Official repository for our paper “Improving black-box optimization in VAE latent space using decoder uncertainty”. The codebase provides an example application in molecular optimization with JT-VAE for both Gradient Ascent and Bayesian Optimization.
CodePascal Notin
Disease variant prediction with deep generative models of evolutionary data
Official repository for our paper “Disease variant prediction with deep generative models of evolutionary data”. The code provides all functionalities required to train the different models leveraged in EVE (Evolutionary model of Variant Effect), as well as the ones to score any mutated protein sequence of interest.
CodePascal Notin
Blog Posts
OATML Conference papers at NeurIPS 2022
OATML group members and collaborators are proud to present 8 papers at NeurIPS 2022 main conference, and 11 workshop papers. …
Full post...Yarin Gal, Freddie Kalaitzis, Shreshth Malik, Lorenz Kuhn, Gunshi Gupta, Jannik Kossen, Pascal Notin, Andrew Jesson, Panagiotis Tigas, Tim G. J. Rudner, Sebastian Farquhar, Ilia Shumailov, 25 Nov 2022
OATML at ICML 2022
OATML group members and collaborators are proud to present 11 papers at the ICML 2022 main conference and workshops. Group members are also co-organizing the Workshop on Computational Biology, and the Oxford Wom*n Social. …
Full post...Sören Mindermann, Jan Brauner, Muhammed Razzak, Andreas Kirsch, Aidan Gomez, Sebastian Farquhar, Pascal Notin, Tim G. J. Rudner, Freddie Bickford Smith, Neil Band, Panagiotis Tigas, Andrew Jesson, Lars Holdijk, Joost van Amersfoort, Kelsey Doerksen, Jannik Kossen, Yarin Gal, 17 Jul 2022
OATML at ICLR 2022
OATML group members and collaborators are proud to present 4 papers at ICLR 2022 main conference. …
Full post...Yarin Gal, Tuan Nguyen, Andrew Jesson, Pascal Notin, Atılım Güneş Baydin, Clare Lyle, Milad Alizadeh, Joost van Amersfoort, Sebastian Farquhar, Muhammed Razzak, Freddie Kalaitzis, 01 Feb 2022
13 OATML Conference papers at NeurIPS 2021
OATML group members and collaborators are proud to present 13 papers at NeurIPS 2021 main conference. …
Full post...Jannik Kossen, Neil Band, Aidan Gomez, Clare Lyle, Tim G. J. Rudner, Yarin Gal, Binxin (Robin) Ru, Clare Lyle, Lisa Schut, Atılım Güneş Baydin, Tim G. J. Rudner, Andrew Jesson, Panagiotis Tigas, Joost van Amersfoort, Andreas Kirsch, Pascal Notin, Angelos Filos, 11 Oct 2021
21 OATML Conference and Workshop papers at ICML 2021
OATML group members and collaborators are proud to present 21 papers at ICML 2021, including 7 papers at the main conference and 14 papers at various workshops. Group members will also be giving invited talks and participate in panel discussions at the workshops. …
Full post...Angelos Filos, Clare Lyle, Jannik Kossen, Sebastian Farquhar, Tom Rainforth, Andrew Jesson, Sören Mindermann, Tim G. J. Rudner, Oscar Key, Binxin (Robin) Ru, Pascal Notin, Panagiotis Tigas, Andreas Kirsch, Jishnu Mukhoti, Joost van Amersfoort, Lisa Schut, Muhammed Razzak, Aidan Gomez, Jan Brauner, Yarin Gal, 17 Jul 2021
13 OATML Conference and Workshop papers at ICML 2020
We are glad to share the following 13 papers by OATML authors and collaborators to be presented at this ICML conference and workshops …
Full post...Angelos Filos, Sebastian Farquhar, Tim G. J. Rudner, Lewis Smith, Lisa Schut, Tom Rainforth, Panagiotis Tigas, Pascal Notin, Andreas Kirsch, Clare Lyle, Joost van Amersfoort, Jishnu Mukhoti, Yarin Gal, 10 Jul 2020